+Search query
-Structure paper
| Title | Shared structural mechanisms of general anaesthetics and benzodiazepines. |
|---|---|
| Journal, issue, pages | Nature, Vol. 585, Issue 7824, Page 303-308, Year 2020 |
| Publish date | Sep 2, 2020 |
 Authors Authors | Jeong Joo Kim / Anant Gharpure / Jinfeng Teng / Yuxuan Zhuang / Rebecca J Howard / Shaotong Zhu / Colleen M Noviello / Richard M Walsh / Erik Lindahl / Ryan E Hibbs /   |
| PubMed Abstract | Most general anaesthetics and classical benzodiazepine drugs act through positive modulation of γ-aminobutyric acid type A (GABA) receptors to dampen neuronal activity in the brain. However, direct ...Most general anaesthetics and classical benzodiazepine drugs act through positive modulation of γ-aminobutyric acid type A (GABA) receptors to dampen neuronal activity in the brain. However, direct structural information on the mechanisms of general anaesthetics at their physiological receptor sites is lacking. Here we present cryo-electron microscopy structures of GABA receptors bound to intravenous anaesthetics, benzodiazepines and inhibitory modulators. These structures were solved in a lipidic environment and are complemented by electrophysiology and molecular dynamics simulations. Structures of GABA receptors in complex with the anaesthetics phenobarbital, etomidate and propofol reveal both distinct and common transmembrane binding sites, which are shared in part by the benzodiazepine drug diazepam. Structures in which GABA receptors are bound by benzodiazepine-site ligands identify an additional membrane binding site for diazepam and suggest an allosteric mechanism for anaesthetic reversal by flumazenil. This study provides a foundation for understanding how pharmacologically diverse and clinically essential drugs act through overlapping and distinct mechanisms to potentiate inhibitory signalling in the brain. |
 External links External links |  Nature / Nature /  PubMed:32879488 / PubMed:32879488 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.55 - 3.5 Å |
| Structure data | EMDB-22031, PDB-6x3s: EMDB-22032, PDB-6x3t: EMDB-22033, PDB-6x3u: EMDB-22034, PDB-6x3v: EMDB-22035, PDB-6x3w: EMDB-22036, PDB-6x3x: EMDB-22037, PDB-6x3z: EMDB-22038, PDB-6x40: |
| Chemicals |  ChemComp-NAG:  ChemComp-J94:  ChemComp-MAN:  ChemComp-ABU: 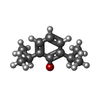 ChemComp-PFL: 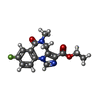 ChemComp-FYP: 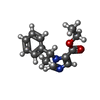 ChemComp-V8D:  ChemComp-UQA: 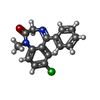 ChemComp-DZP:  ChemComp-RI5: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / Ion channel / Cys-loop receptor / pentametic ligand gated channel / GABAA receptor / TRANSPORT PROTEIN / pentametic ligand gated ion channel |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



















 homo sapiens (human)
homo sapiens (human)
