+Search query
-Structure paper
| Title | Exit tunnel modulation as resistance mechanism of S. aureus erythromycin resistant mutant. |
|---|---|
| Journal, issue, pages | Sci Rep, Vol. 9, Issue 1, Page 11460, Year 2019 |
| Publish date | Aug 7, 2019 |
 Authors Authors | Yehuda Halfon / Donna Matzov / Zohar Eyal / Anat Bashan / Ella Zimmerman / Jette Kjeldgaard / Hanne Ingmer / Ada Yonath /   |
| PubMed Abstract | The clinical use of the antibiotic erythromycin (ery) is hampered owing to the spread of resistance genes that are mostly mutating rRNA around the ery binding site at the entrance to the protein exit ...The clinical use of the antibiotic erythromycin (ery) is hampered owing to the spread of resistance genes that are mostly mutating rRNA around the ery binding site at the entrance to the protein exit tunnel. Additional effective resistance mechanisms include deletion or insertion mutations in ribosomal protein uL22, which lead to alterations of the exit tunnel shape, located 16 Å away from the drug's binding site. We determined the cryo-EM structures of the Staphylococcus aureus 70S ribosome, and its ery bound complex with a two amino acid deletion mutation in its ß hairpin loop, which grants the bacteria resistance to ery. The structures reveal that, although the binding of ery is stable, the movement of the flexible shorter uL22 loop towards the tunnel wall creates a wider path for nascent proteins, thus enabling bypass of the barrier formed by the drug. Moreover, upon drug binding, the tunnel widens further. |
 External links External links |  Sci Rep / Sci Rep /  PubMed:31391518 / PubMed:31391518 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.3 - 3.58 Å |
| Structure data | EMDB-10076, PDB-6s0x: EMDB-10077, PDB-6s0z: EMDB-10078, PDB-6s12: EMDB-10079, PDB-6s13: |
| Chemicals | 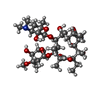 ChemComp-ERY: |
| Source |
|
 Keywords Keywords | RIBOSOME / antibiotics / resistance / Staphylococcus aureus / exit tunnel / RNA / rProteins / erythromycin |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers












