+Search query
-Structure paper
| Title | Development of a Novel Lead that Targets M. tuberculosis Polyketide Synthase 13. |
|---|---|
| Journal, issue, pages | Cell, Vol. 170, Page 249-259.e25, Year 2017 |
| Publish date | Mar 8, 2017 (structure data deposition date) |
 Authors Authors | Aggarwal, A. / Parai, M.K. / Shetty, N. / Wallis, D. / Woolhiser, L. / Hastings, C. / Dutta, N.K. / Galaviz, S. / Dhakal, R.C. / Shrestha, R. ...Aggarwal, A. / Parai, M.K. / Shetty, N. / Wallis, D. / Woolhiser, L. / Hastings, C. / Dutta, N.K. / Galaviz, S. / Dhakal, R.C. / Shrestha, R. / Wakabayashi, S. / Walpole, C. / Matthews, D. / Floyd, D. / Scullion, P. / Riley, J. / Epemolu, O. / Norval, S. / Snavely, T. / Robertson, G.T. / Rubin, E.J. / Ioerger, T.R. / Sirgel, F.A. / van der Merwe, R. / van Helden, P.D. / Keller, P. / Bottger, E.C. / Karakousis, P.C. / Lenaerts, A.J. / Sacchettini, J.C. |
 External links External links |  Cell / Cell /  PubMed:28669536 PubMed:28669536 |
| Methods | X-ray diffraction |
| Resolution | 1.723 - 2.051 Å |
| Structure data |  PDB-5v3w:  PDB-5v3x:  PDB-5v3y:  PDB-5v3z:  PDB-5v40:  PDB-5v41:  PDB-5v42: |
| Chemicals | 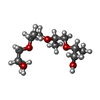 ChemComp-2Q5:  ChemComp-HOH: 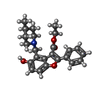 ChemComp-I28: 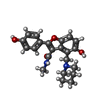 ChemComp-5V8:  ChemComp-SO4:  ChemComp-1PE: 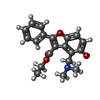 ChemComp-JS1:  ChemComp-J24:  ChemComp-I66: |
| Source |
|
 Keywords Keywords | TRANSFERASE / thioesterase domain / Pks13 / Mycobacterium / polyketide synthase / mycolic acid condensation / TB Structural Genomics Consortium / TBSGC / alpha/beta hydrolase / thioesterase / TRANSFERASE/TRANSFERASE INHIBITOR / TAM1 complex / TRANSFERASE-TRANSFERASE INHIBITOR complex / TAM16 complex / THIOESTERASE-TRANSFERASE INHIBITOR complex / D1607N mutant / TAM6 complex / TAM5 complex / TAM3 complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




