+Search query
-Structure paper
| Title | De novo modeling of the F(420)-reducing [NiFe]-hydrogenase from a methanogenic archaeon by cryo-electron microscopy. |
|---|---|
| Journal, issue, pages | Elife, Vol. 2, Page e00218, Year 2013 |
| Publish date | Mar 5, 2013 |
 Authors Authors | Deryck J Mills / Stella Vitt / Mike Strauss / Seigo Shima / Janet Vonck /  |
| PubMed Abstract | Methanogenic archaea use a [NiFe]-hydrogenase, Frh, for oxidation/reduction of F420, an important hydride carrier in the methanogenesis pathway from H2 and CO2. Frh accounts for about 1% of the ...Methanogenic archaea use a [NiFe]-hydrogenase, Frh, for oxidation/reduction of F420, an important hydride carrier in the methanogenesis pathway from H2 and CO2. Frh accounts for about 1% of the cytoplasmic protein and forms a huge complex consisting of FrhABG heterotrimers with each a [NiFe] center, four Fe-S clusters and an FAD. Here, we report the structure determined by near-atomic resolution cryo-EM of Frh with and without bound substrate F420. The polypeptide chains of FrhB, for which there was no homolog, was traced de novo from the EM map. The 1.2-MDa complex contains 12 copies of the heterotrimer, which unexpectedly form a spherical protein shell with a hollow core. The cryo-EM map reveals strong electron density of the chains of metal clusters running parallel to the protein shell, and the F420-binding site is located at the end of the chain near the outside of the spherical structure. DOI:http://dx.doi.org/10.7554/eLife.00218.001. |
 External links External links |  Elife / Elife /  PubMed:23483797 / PubMed:23483797 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 4 - 4.5 Å |
| Structure data | |
| Chemicals |  ChemComp-FCO:  ChemComp-NI:  ChemComp-FE2:  ChemComp-SF4: 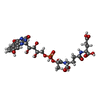 ChemComp-F42:  ChemComp-FAD: |
| Source |
|
 Keywords Keywords | OXIDOREDUCTASE / METHANOGENESIS |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers







 methanothermobacter marburgensis (archaea)
methanothermobacter marburgensis (archaea)