+Search query
-Structure paper
| Title | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase |
|---|---|
| Journal, issue, pages | J. Biol. Chem., Vol. 284, Page 27637-27645, Year 2009 |
| Publish date | Jul 12, 2009 (structure data deposition date) |
 Authors Authors | Liu, Q. / Graeff, R. / Kriksunov, I.A. / Jiang, H. / Zhang, B. / Oppenheimer, N. / Lin, H. / Potter, B.V.L. / Lee, H.C. / Hao, Q. |
 External links External links |  J. Biol. Chem. / J. Biol. Chem. /  PubMed:19640846 PubMed:19640846 |
| Methods | X-ray diffraction |
| Resolution | 1.75 - 2.18 Å |
| Structure data |  PDB-3i9j:  PDB-3i9k:  PDB-3i9l:  PDB-3i9m:  PDB-3i9n: |
| Chemicals |  ChemComp-NFD: 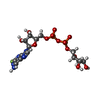 ChemComp-AVV: 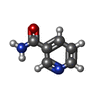 ChemComp-NCA:  ChemComp-HOH:  ChemComp-NAD:  ChemComp-N1C:  ChemComp-AVU:  ChemComp-AVW: |
| Source |
|
 Keywords Keywords | HYDROLASE / Homodimer / enzyme-analog-nicotinamide complex / ADP-ribosyl cyclase / Disulfide bond / Fertilization / NAD / protein substrate NAD complex / enzyme-product analog complex / Enzyme-analog complex / covalent reaction intermediate / Alpha helices rich domain and alpha/beta domain / Alternative splicing / Diabetes mellitus / Glycoprotein / Membrane / Polymorphism / Receptor / Signal-anchor / Transmembrane |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




 homo sapiens (human)
homo sapiens (human)