+検索条件
-Structure paper
| タイトル | Structural basis of human ABCC4 recognition of cAMP and ligand recognition flexibility. |
|---|---|
| ジャーナル・号・ページ | Cell Biosci, Vol. 15, Issue 1, Page 39, Year 2025 |
| 掲載日 | 2025年3月27日 |
 著者 著者 | Xuepeng Wen / Kaixue Si / Dantong Zhu / Anqi Zhang / Changyou Guo / Minghui Li / Weiming Tian /  |
| PubMed 要旨 | BACKGROUND: ABCC4 (ATP-binding cassette sub-family C member 4) is a transporter protein that is primarily localized to the plasma membrane, and its efflux activity is associated with the progression ...BACKGROUND: ABCC4 (ATP-binding cassette sub-family C member 4) is a transporter protein that is primarily localized to the plasma membrane, and its efflux activity is associated with the progression of various cancers and the development of drug resistance. Cyclic adenosine monophosphate (cAMP) is an important biomolecule that is considered a transport substrate of ABCC4. However, there is currently no direct structural understanding of how ABCC4 binds cAMP, and the mechanisms by which it recognizes a diverse range of substrate ligands remain poorly understood. Some studies have indicated that, under physiological conditions, cAMP does not significantly stimulate the ATPase activity of ABCC4, making the commonly used ATPase activity assays for ABC proteins unsuitable for studying cAMP. RESULTS: Here, we successfully resolved the cryo-electron microscopy (cryo-EM) structure of the human ABCC4-cAMP (hABCC4-cAMP) complex, revealing how hABCC4 binds to cAMP and identifying the key residues involved. This structure was compared with two other hABCC4 complex structures we obtained (Methotrexate and Prostaglandin E) and with previously published structures. We discovered some new structural insights into how hABCC4 binds ligands. On the basis of the structural information obtained, we confirmed the feasibility of using 8-[Fluo]-cAMP in a transport assay to detect cAMP translocation and found that some challenges remain to be addressed. CONCLUSIONS: These results suggest that hABCC4 can bind cAMP and exhibits varying degrees of flexibility when binding with different substrates, including cAMP. These findings expand our understanding of the structural biology of ABCC4. |
 リンク リンク |  Cell Biosci / Cell Biosci /  PubMed:40148998 / PubMed:40148998 /  PubMed Central PubMed Central |
| 手法 | EM (単粒子) |
| 解像度 | 2.99 - 3.29 Å |
| 構造データ | EMDB-62531, PDB-9krk: EMDB-62532, PDB-9krl: EMDB-62533, PDB-9krm: EMDB-62534, PDB-9krn: |
| 化合物 |  ChemComp-CMP: 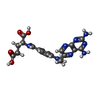 ChemComp-MTX: 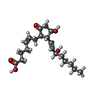 ChemComp-P2E: |
| 由来 |
|
 キーワード キーワード | TRANSPORT PROTEIN / ABC-type transporter activity / ATP hydrolysis activity / ATPase-coupled transmembrane transporter activity / ATP binding / Transport protein. |
 ムービー
ムービー コントローラー
コントローラー 構造ビューア
構造ビューア 万見文献について
万見文献について











 homo sapiens (ヒト)
homo sapiens (ヒト)