+Search query
-Structure paper
| Title | Molecular mechanisms of CBASS phospholipase effector CapV mediated membrane disruption. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 16, Issue 1, Page 8611, Year 2025 |
| Publish date | Sep 29, 2025 |
 Authors Authors | Jianping Kong / Wanqian Wu / Shiyue Ke / Zihan Zhou / Shenglan Xia / Jianyu Chen / Runyu Zhu / Yijia Hou / Tinashe Makanyire / Xiangru Shan / Zhuyue Zhuo / Keying Li / Hongtao Shen / Pan Yang / Pingping Huang / Jingxian Liu / Jing Li / Xiaolian Sun / Jiajia Dong / Hongbin Sun / Meirong Chen / Meiling Lu / Zhaoxing Li / Yibei Xiao /  |
| PubMed Abstract | Cyclic oligonucleotide-based antiphage signaling systems (CBASS) are widespread bacterial immune systems that trigger host suicide via cyclic nucleotide-activated effectors. The predominant strategy ...Cyclic oligonucleotide-based antiphage signaling systems (CBASS) are widespread bacterial immune systems that trigger host suicide via cyclic nucleotide-activated effectors. The predominant strategy to induce cell death in CBASS is membrane disruption. Here, we demonstrate that patatin-like phospholipase CapV, the most abundant CBASS effector, relocates and cleaves membrane phospholipids at the cell pole upon 3'3'-cGAMP binding, inducing polarized membrane disruption and cell death. Using cryo-EM, we reveal that apo-CapV adopts both dimeric and tetrameric states, with its phospholipid-binding pocket occluded and locked in an inactive conformation. Binding to 3'3'-cGAMP induces filamentation and substantial conformational change of CapV, enhancing membrane binding via electrostatic interactions between its interspaced basic surfaces and the negatively charged phosphate moieties of phospholipids. Simultaneously, the rearrangement opens the phospholipid-binding pocket, enabling the accommodation of two fatty acid chains of phospholipid within distinct hydrophobic pockets. Our findings reveal a filament-dependent activation mechanism for phospholipase-mediated membrane disruption during antiviral response. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:41022836 / PubMed:41022836 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.7 - 3.1 Å |
| Structure data | EMDB-60393: Cryo-EM structure of AbCapV filemant bound with 3',3'-cGAMP with extra phospholipid density EMDB-61417, PDB-9jeh: EMDB-61419, PDB-9jek: EMDB-62291: Cryo-EM structure of AbCapV S58A filament bound with 3'3'-cGAMP with extra phospholipid density |
| Chemicals | 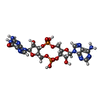 ChemComp-4BW: |
| Source |
|
 Keywords Keywords | HYDROLASE / CBASS / phospholipase |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers











 acinetobacter baumannii (bacteria)
acinetobacter baumannii (bacteria)