[English] 日本語
 Yorodumi
Yorodumi- EMDB-62291: Cryo-EM structure of AbCapV S58A filament bound with 3'3'-cGAMP w... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of AbCapV S58A filament bound with 3'3'-cGAMP with extra phospholipid density | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CBASS / phospholipase / HYDROLASE | |||||||||
| Function / homology | Patatin-like phospholipase domain / Patatin-like phospholipase / Patatin-like phospholipase (PNPLA) domain profile. / Acyl transferase/acyl hydrolase/lysophospholipase / lipid catabolic process / hydrolase activity / CGAMP-activated phospholipase CapV Function and homology information Function and homology information | |||||||||
| Biological species |  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Kong JP / Li ZX / Wu WQ / Xiao YB | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Molecular mechanisms of CBASS phospholipase effector CapV mediated membrane disruption. Authors: Jianping Kong / Wanqian Wu / Shiyue Ke / Zihan Zhou / Shenglan Xia / Jianyu Chen / Runyu Zhu / Yijia Hou / Tinashe Makanyire / Xiangru Shan / Zhuyue Zhuo / Keying Li / Hongtao Shen / Pan ...Authors: Jianping Kong / Wanqian Wu / Shiyue Ke / Zihan Zhou / Shenglan Xia / Jianyu Chen / Runyu Zhu / Yijia Hou / Tinashe Makanyire / Xiangru Shan / Zhuyue Zhuo / Keying Li / Hongtao Shen / Pan Yang / Pingping Huang / Jingxian Liu / Jing Li / Xiaolian Sun / Jiajia Dong / Hongbin Sun / Meirong Chen / Meiling Lu / Zhaoxing Li / Yibei Xiao /  Abstract: Cyclic oligonucleotide-based antiphage signaling systems (CBASS) are widespread bacterial immune systems that trigger host suicide via cyclic nucleotide-activated effectors. The predominant strategy ...Cyclic oligonucleotide-based antiphage signaling systems (CBASS) are widespread bacterial immune systems that trigger host suicide via cyclic nucleotide-activated effectors. The predominant strategy to induce cell death in CBASS is membrane disruption. Here, we demonstrate that patatin-like phospholipase CapV, the most abundant CBASS effector, relocates and cleaves membrane phospholipids at the cell pole upon 3'3'-cGAMP binding, inducing polarized membrane disruption and cell death. Using cryo-EM, we reveal that apo-CapV adopts both dimeric and tetrameric states, with its phospholipid-binding pocket occluded and locked in an inactive conformation. Binding to 3'3'-cGAMP induces filamentation and substantial conformational change of CapV, enhancing membrane binding via electrostatic interactions between its interspaced basic surfaces and the negatively charged phosphate moieties of phospholipids. Simultaneously, the rearrangement opens the phospholipid-binding pocket, enabling the accommodation of two fatty acid chains of phospholipid within distinct hydrophobic pockets. Our findings reveal a filament-dependent activation mechanism for phospholipase-mediated membrane disruption during antiviral response. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_62291.map.gz emd_62291.map.gz | 32.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-62291-v30.xml emd-62291-v30.xml emd-62291.xml emd-62291.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_62291_fsc.xml emd_62291_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_62291.png emd_62291.png | 105.4 KB | ||
| Filedesc metadata |  emd-62291.cif.gz emd-62291.cif.gz | 6.3 KB | ||
| Others |  emd_62291_additional_1.map.gz emd_62291_additional_1.map.gz emd_62291_half_map_1.map.gz emd_62291_half_map_1.map.gz emd_62291_half_map_2.map.gz emd_62291_half_map_2.map.gz | 59.5 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-62291 http://ftp.pdbj.org/pub/emdb/structures/EMD-62291 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-62291 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-62291 | HTTPS FTP |
-Related structure data
| Related structure data |  9kejMC  8zr9C  9jehC  9jekC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_62291.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_62291.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.932 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_62291_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_62291_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_62291_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : AbCapV S58A with 3'3'-cGAMP
| Entire | Name: AbCapV S58A with 3'3'-cGAMP |
|---|---|
| Components |
|
-Supramolecule #1: AbCapV S58A with 3'3'-cGAMP
| Supramolecule | Name: AbCapV S58A with 3'3'-cGAMP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
-Macromolecule #1: CGAMP-activated phospholipase CapV
| Macromolecule | Name: CGAMP-activated phospholipase CapV / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
| Molecular weight | Theoretical: 41.291309 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: METSENKSEI KILSLNGGGV RGLFTITLLA ELESIIEKRE KCENVKIGDY FDLITGTAIG GILALGLASG KSARELKEAF EINATKIFP LKRFKNKQWW NLLRRSIYES EPLYDAVKSM IGETIKFEDL NRRVMITSVN LSTGKPKFFK TPHNPMFTMD R EIRLIDAA ...String: METSENKSEI KILSLNGGGV RGLFTITLLA ELESIIEKRE KCENVKIGDY FDLITGTAIG GILALGLASG KSARELKEAF EINATKIFP LKRFKNKQWW NLLRRSIYES EPLYDAVKSM IGETIKFEDL NRRVMITSVN LSTGKPKFFK TPHNPMFTMD R EIRLIDAA MATSAAPTYF KPHYIEKLEN YFADGGLVAN NPSYIGIREV LIDMKNDFPD AKPENIKVLN IGTLSEDYCI SP ETLSKNS GKGYLSLWNM GERIVLSTMT ANQHLQRFML LREFEALKIE KNYVEIDETI PNEAAAEITL DNASEGCLKA LRG SGKKLA AERYTKNEEL RNFFLKKAEP FVPYIESSEV TAHHHHHH UniProtKB: CGAMP-activated phospholipase CapV |
-Macromolecule #2: 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-...
| Macromolecule | Name: 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2- ...Name: 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one type: ligand / ID: 2 / Number of copies: 6 / Formula: 4BW |
|---|---|
| Molecular weight | Theoretical: 674.411 Da |
| Chemical component information | 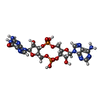 ChemComp-4BW: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































