+Search query
-Structure paper
| Title | Cryo-EM structures of ryanodine receptors and diamide insecticides reveal the mechanisms of selectivity and resistance. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 15, Issue 1, Page 9056, Year 2024 |
| Publish date | Oct 20, 2024 |
 Authors Authors | Lianyun Lin / Changshi Wang / Wenlan Wang / Heng Jiang / Takashi Murayama / Takuya Kobayashi / Hadiatullah Hadiatullah / Yu Seby Chen / Shunfan Wu / Yiwen Wang / Henryk Korza / Yucheng Gu / Yan Zhang / Jiamu Du / Filip Van Petegem / Zhiguang Yuchi /     |
| PubMed Abstract | The resistance of pests to common insecticides is a global issue that threatens food production worldwide. Diamide insecticides target insect ryanodine receptors (RyRs), causing uncontrolled calcium ...The resistance of pests to common insecticides is a global issue that threatens food production worldwide. Diamide insecticides target insect ryanodine receptors (RyRs), causing uncontrolled calcium release from the sarcoplasmic and endoplasmic reticulum. Despite their high potency and species selectivity, several resistance mutations have emerged. Using a chimeric RyR (chiRyR) approach and cryo-electron microscopy (cryo-EM), we investigate how insect RyRs engage two different diamide insecticides from separate families: flubendiamide, a phthalic acid derivative, and tetraniliprole, an anthranilic compound. Both compounds target the same site in the transmembrane region of the RyR, albeit with different poses, and promote channel opening through coupling with the pore-forming domain. To explore the resistance mechanisms, we also solve two cryo-EM structures of chiRyR carrying the two most common resistance mutations, I4790M and G4946E, both alone and in complex with the diamide insecticide chlorantraniliprole. The resistance mutations perturb the local structure, directly reducing the binding affinity and altering the binding pose. Our findings elucidate the mode of action of different diamide insecticides, reveal the molecular mechanism of resistance mutations, and provide important clues for the development of novel pesticides that can bypass the resistance mutations. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:39428398 / PubMed:39428398 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.28 - 3.91 Å |
| Structure data | EMDB-38398, PDB-8xji: EMDB-38417, PDB-8xkh: EMDB-38447, PDB-8xlf: EMDB-38448, PDB-8xlh:  EMDB-38551: The map of chimeric RyR transmembrane domain in complex with flubendiamide after TMD local refinement  EMDB-38553: The map of chimeric RyR transmembrane domain in complex with tetraniliprole with TMD local refinement EMDB-38908, PDB-8y40:  EMDB-60899: Structure of a chimeric RyR-I4657M/G4819E (local refinement of TMD)  EMDB-60900: Cryo-EM structure of ref-chiRyR (local refinement of TMD)  EMDB-60901: cryo-EM structure of chiRyR-I4657M/G4819E complex with CHL (local refinement of TMD) |
| Chemicals |  PDB-1lvx:  ChemComp-ZN:  ChemComp-CA:  ChemComp-ATP: 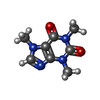 ChemComp-CFF:  PDB-1lv1: 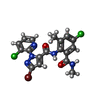 ChemComp-F0U: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / Ryanodine receptor / Ion channel / tetraniliprole / chlorantraniliprole |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers














 homo sapiens (human)
homo sapiens (human)