+Search query
-Structure paper
| Title | Characterization of a broadly neutralizing monoclonal antibody that targets the fusion domain of group 2 influenza A virus hemagglutinin. |
|---|---|
| Journal, issue, pages | J Virol, Vol. 88, Issue 23, Page 13580-13592, Year 2014 |
| Publish date | Sep 10, 2014 |
 Authors Authors | Gene S Tan / Peter S Lee / Ryan M B Hoffman / Beryl Mazel-Sanchez / Florian Krammer / Paul E Leon / Andrew B Ward / Ian A Wilson / Peter Palese /  |
| PubMed Abstract | Due to continuous changes to its antigenic regions, influenza viruses can evade immune detection and cause a significant amount of morbidity and mortality around the world. Influenza vaccinations can ...Due to continuous changes to its antigenic regions, influenza viruses can evade immune detection and cause a significant amount of morbidity and mortality around the world. Influenza vaccinations can protect against disease but must be annually reformulated to match the current circulating strains. In the development of a broad-spectrum influenza vaccine, the elucidation of conserved epitopes is paramount. To this end, we designed an immunization strategy in mice to boost the humoral response against conserved regions of the hemagglutinin (HA) glycoprotein. Of note, generation and identification of broadly neutralizing antibodies that target group 2 HAs are rare and thus far have yielded only a few monoclonal antibodies (MAbs). Here, we demonstrate that mouse MAb 9H10 has broad and potent in vitro neutralizing activity against H3 and H10 group 2 influenza A subtypes. In the mouse model, MAb 9H10 protects mice against two divergent mouse-adapted H3N2 strains, in both pre- and postexposure administration regimens. In vitro and cell-free assays suggest that MAb 9H10 inhibits viral replication by blocking HA-dependent fusion of the viral and endosomal membranes early in the replication cycle and by disrupting viral particle egress in the late stage of infection. Interestingly, electron microscopy reconstructions of MAb 9H10 bound to the HA reveal that it binds a similar binding footprint to MAbs CR8020 and CR8043. IMPORTANCE: The influenza hemagglutinin is the major antigenic target of the humoral immune response. However, due to continuous antigenic changes that occur on the surface of this glycoprotein, influenza viruses can escape the immune system and cause significant disease to the host. Toward the development of broad-spectrum therapeutics and vaccines against influenza virus, elucidation of conserved regions of influenza viruses is crucial. Thus, defining these types of epitopes through the generation and characterization of broadly neutralizing monoclonal antibodies (MAbs) can greatly assist others in highlighting conserved regions of hemagglutinin. Here, we demonstrate that MAb 9H10 that targets the hemagglutinin stalk has broadly neutralizing activity against group 2 influenza A viruses in vitro and in vivo. |
 External links External links |  J Virol / J Virol /  PubMed:25210195 / PubMed:25210195 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 24.6 Å |
| Structure data | 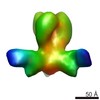 EMDB-6077: |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




 Influenza A virus
Influenza A virus
