+Search query
-Structure paper
| Title | Discovery and structural basis of endogenous and exogenous inhibitors of the proton-activated-chloride channel. |
|---|---|
| Journal, issue, pages | Cell Rep, Vol. 44, Issue 7, Page 115998, Year 2025 |
| Publish date | Jul 22, 2025 |
 Authors Authors | Heng Zhang / Zhijun Zhao / Xiaoying Chen / Qingxia Ma / Wenxuan Zhen / Qianqian Xu / Yaqi Wang / Bingyu He / Ying Huang / Pengfei Xu / Haoxing Xu / Zhimin Lu / Nannan Su / Fan Yang /  |
| PubMed Abstract | The proton-activated chloride (PAC) channel is a broadly expressed chloride channel that senses extracellular acidification, regulates endosomal acidification, and participates in many processes, ...The proton-activated chloride (PAC) channel is a broadly expressed chloride channel that senses extracellular acidification, regulates endosomal acidification, and participates in many processes, including acid-induced cell death and cancer cell migration. However, the lack of specific modulators has limited our understanding of its function and therapeutic potential. Here, we discovered that endogenous cholesterol (CHL) partially inhibits PAC channel activation. Moreover, we identified a series of CHL analogs and structurally related dyes as full antagonists of the PAC channel, with deoxycholic acid (DCA) and Evans blue (EB) as their representative compounds. By combining cryo-electron microscopy (cryo-EM) and patch-clamp recordings, we revealed the distinct binding and inhibition mechanisms of DCA and EB on this channel. Moreover, in malignant osteosarcoma cells, where PAC channel expression is upregulated, EB inhibited the migration and invasion of these cells. Therefore, we identified DCA and EB as pharmacological tools for the PAC channel, which forms a basis for future drug discovery targeting this channel. |
 External links External links |  Cell Rep / Cell Rep /  PubMed:40664207 PubMed:40664207 |
| Methods | EM (single particle) |
| Resolution | 2.89 - 3.85 Å |
| Structure data | EMDB-60224, PDB-8zlb: EMDB-60228, PDB-8zll: EMDB-63102, PDB-9lhm: EMDB-63110, PDB-9li2: |
| Chemicals | 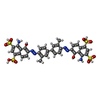 ChemComp-IZ8:  ChemComp-CLR: 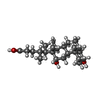 ChemComp-DXC: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / hPAC / EVANS BLUE / STRUCTUAL PROTEIN / PAC / DCA |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers











 homo sapiens (human)
homo sapiens (human)