+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | EB bound state of hPAC | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | hPAC / EVANS BLUE / STRUCTUAL PROTEIN / MEMBRANE PROTEIN | |||||||||
| Function / homology | pH-gated chloride channel activity / TMEM206 protein / TMEM206 protein family / chloride transport / chloride channel complex / cell surface / plasma membrane / Proton-activated chloride channel Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.85 Å | |||||||||
 Authors Authors | Su N / Chen X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2025 Journal: Cell Rep / Year: 2025Title: Discovery and structural basis of endogenous and exogenous inhibitors of the proton-activated-chloride channel. Authors: Heng Zhang / Zhijun Zhao / Xiaoying Chen / Qingxia Ma / Wenxuan Zhen / Qianqian Xu / Yaqi Wang / Bingyu He / Ying Huang / Pengfei Xu / Haoxing Xu / Zhimin Lu / Nannan Su / Fan Yang /  Abstract: The proton-activated chloride (PAC) channel is a broadly expressed chloride channel that senses extracellular acidification, regulates endosomal acidification, and participates in many processes, ...The proton-activated chloride (PAC) channel is a broadly expressed chloride channel that senses extracellular acidification, regulates endosomal acidification, and participates in many processes, including acid-induced cell death and cancer cell migration. However, the lack of specific modulators has limited our understanding of its function and therapeutic potential. Here, we discovered that endogenous cholesterol (CHL) partially inhibits PAC channel activation. Moreover, we identified a series of CHL analogs and structurally related dyes as full antagonists of the PAC channel, with deoxycholic acid (DCA) and Evans blue (EB) as their representative compounds. By combining cryo-electron microscopy (cryo-EM) and patch-clamp recordings, we revealed the distinct binding and inhibition mechanisms of DCA and EB on this channel. Moreover, in malignant osteosarcoma cells, where PAC channel expression is upregulated, EB inhibited the migration and invasion of these cells. Therefore, we identified DCA and EB as pharmacological tools for the PAC channel, which forms a basis for future drug discovery targeting this channel. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60224.map.gz emd_60224.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60224-v30.xml emd-60224-v30.xml emd-60224.xml emd-60224.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_60224_fsc.xml emd_60224_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_60224.png emd_60224.png | 43.2 KB | ||
| Filedesc metadata |  emd-60224.cif.gz emd-60224.cif.gz | 5.8 KB | ||
| Others |  emd_60224_half_map_1.map.gz emd_60224_half_map_1.map.gz emd_60224_half_map_2.map.gz emd_60224_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60224 http://ftp.pdbj.org/pub/emdb/structures/EMD-60224 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60224 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60224 | HTTPS FTP |
-Validation report
| Summary document |  emd_60224_validation.pdf.gz emd_60224_validation.pdf.gz | 893.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60224_full_validation.pdf.gz emd_60224_full_validation.pdf.gz | 893.5 KB | Display | |
| Data in XML |  emd_60224_validation.xml.gz emd_60224_validation.xml.gz | 15.9 KB | Display | |
| Data in CIF |  emd_60224_validation.cif.gz emd_60224_validation.cif.gz | 21 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60224 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60224 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60224 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60224 | HTTPS FTP |
-Related structure data
| Related structure data |  8zlbMC  8zllC  9lhmC  9li2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_60224.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60224.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.014 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_60224_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_60224_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of hPAC and EB
| Entire | Name: Complex of hPAC and EB |
|---|---|
| Components |
|
-Supramolecule #1: Complex of hPAC and EB
| Supramolecule | Name: Complex of hPAC and EB / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Proton-activated chloride channel
| Macromolecule | Name: Proton-activated chloride channel / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 44.681938 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDDDDKW SHPQFEKGGG GSGGSAWSHP QFEKEFKGLV DMIRQERSTS YQELSEELVQ VVENSELADE QDKETVRVQG PGILPGLDS ESASSSIRFS KACLKNVFSV LLIFIYLLLM AVAVFLVYRT ITDFREKLKH PVMSVSYKEV DRYDAPGIAL Y PGQAQLLS ...String: MDYKDDDDKW SHPQFEKGGG GSGGSAWSHP QFEKEFKGLV DMIRQERSTS YQELSEELVQ VVENSELADE QDKETVRVQG PGILPGLDS ESASSSIRFS KACLKNVFSV LLIFIYLLLM AVAVFLVYRT ITDFREKLKH PVMSVSYKEV DRYDAPGIAL Y PGQAQLLS CKHHYEVIPP LTSPGQPGDM NCTTQRINYT DPFSNQTVKS ALIVQGPREV KKRELVFLQF RLNKSSEDFS AI DYLLFSS FQEFLQSPNR VGFMQACESA YSSWKFSGGF RTWVKMSLVK TKEEDGREAV EFRQETSVVN YIDQRPAAKK SAQ LFFVVF EWKDPFIQKV QDIVTANPWN TIALLCGAFL ALFKAAEFAK LSIKWMIKIR KRYLKRRGQA TSHIS UniProtKB: Proton-activated chloride channel |
-Macromolecule #2: 4-azanyl-6-[[4-[4-[(~{E})-(8-azanyl-1-oxidanyl-5,7-disulfo-naphth...
| Macromolecule | Name: 4-azanyl-6-[[4-[4-[(~{E})-(8-azanyl-1-oxidanyl-5,7-disulfo-naphthalen-2-yl)diazenyl]-3-methyl-phenyl]-2-methyl-phenyl]diazenyl]-5-oxidanyl-naphthalene-1,3-disulfonic acid type: ligand / ID: 2 / Number of copies: 1 / Formula: IZ8 |
|---|---|
| Molecular weight | Theoretical: 872.878 Da |
| Chemical component information | 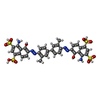 ChemComp-IZ8: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average exposure time: 8.0 sec. / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































