+Search query
-Structure paper
| Title | Structural Basis of Transcription Inhibition by Fidaxomicin (Lipiarmycin A3). |
|---|---|
| Journal, issue, pages | Mol Cell, Vol. 70, Issue 1, Page 60-71.e15, Year 2018 |
| Publish date | Apr 5, 2018 |
 Authors Authors | Wei Lin / Kalyan Das / David Degen / Abhishek Mazumder / Diego Duchi / Dongye Wang / Yon W Ebright / Richard Y Ebright / Elena Sineva / Matthew Gigliotti / Aashish Srivastava / Sukhendu Mandal / Yi Jiang / Yu Liu / Ruiheng Yin / Zhening Zhang / Edward T Eng / Dennis Thomas / Stefano Donadio / Haibo Zhang / Changsheng Zhang / Achillefs N Kapanidis / Richard H Ebright /      |
| PubMed Abstract | Fidaxomicin is an antibacterial drug in clinical use for treatment of Clostridium difficile diarrhea. The active ingredient of fidaxomicin, lipiarmycin A3 (Lpm), functions by inhibiting bacterial ...Fidaxomicin is an antibacterial drug in clinical use for treatment of Clostridium difficile diarrhea. The active ingredient of fidaxomicin, lipiarmycin A3 (Lpm), functions by inhibiting bacterial RNA polymerase (RNAP). Here we report a cryo-EM structure of Mycobacterium tuberculosis RNAP holoenzyme in complex with Lpm at 3.5-Å resolution. The structure shows that Lpm binds at the base of the RNAP "clamp." The structure exhibits an open conformation of the RNAP clamp, suggesting that Lpm traps an open-clamp state. Single-molecule fluorescence resonance energy transfer experiments confirm that Lpm traps an open-clamp state and define effects of Lpm on clamp dynamics. We suggest that Lpm inhibits transcription by trapping an open-clamp state, preventing simultaneous interaction with promoter -10 and -35 elements. The results account for the absence of cross-resistance between Lpm and other RNAP inhibitors, account for structure-activity relationships of Lpm derivatives, and enable structure-based design of improved Lpm derivatives. |
 External links External links |  Mol Cell / Mol Cell /  PubMed:29606590 / PubMed:29606590 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 3.52 - 3.8 Å |
| Structure data | EMDB-4230, PDB-6fbv:  PDB-6asg: |
| Chemicals |  ChemComp-ZN:  ChemComp-MG: 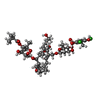 ChemComp-FI8:  ChemComp-HOH: |
| Source |
|
 Keywords Keywords | TRANSCRIPTION / Thermus thermophilus / RNA polymerase core enzyme / Lipiarmycin / RNA pol / RNAP / inhibitor / drug / Clostridium difficile / ANTIBIOTIC / Tiacumicin B / CCDC 114782 |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers





 mycobacterium tuberculosis (strain atcc 25618 / h37rv) (bacteria)
mycobacterium tuberculosis (strain atcc 25618 / h37rv) (bacteria) thermus thermophilus (strain hb8 / atcc 27634 / dsm 579) (bacteria)
thermus thermophilus (strain hb8 / atcc 27634 / dsm 579) (bacteria)