+Search query
-Structure paper
| Title | Structural mechanism of SGLT1 inhibitors. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 13, Issue 1, Page 6440, Year 2022 |
| Publish date | Oct 28, 2022 |
 Authors Authors | Yange Niu / Wenhao Cui / Rui Liu / Sanshan Wang / Han Ke / Xiaoguang Lei / Lei Chen /  |
| PubMed Abstract | Sodium glucose co-transporters (SGLT) harness the electrochemical gradient of sodium to drive the uphill transport of glucose across the plasma membrane. Human SGLT1 (hSGLT1) plays a key role in ...Sodium glucose co-transporters (SGLT) harness the electrochemical gradient of sodium to drive the uphill transport of glucose across the plasma membrane. Human SGLT1 (hSGLT1) plays a key role in sugar uptake from food and its inhibitors show promise in the treatment of several diseases. However, the inhibition mechanism for hSGLT1 remains elusive. Here, we present the cryo-EM structure of the hSGLT1-MAP17 hetero-dimeric complex in the presence of the high-affinity inhibitor LX2761. LX2761 locks the transporter in an outward-open conformation by wedging inside the substrate-binding site and the extracellular vestibule of hSGLT1. LX2761 blocks the putative water permeation pathway of hSGLT1. The structure also uncovers the conformational changes of hSGLT1 during transitions from outward-open to inward-open states. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:36307403 / PubMed:36307403 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.2 Å |
| Structure data | EMDB-32617, PDB-7wmv: |
| Chemicals | 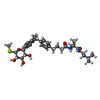 ChemComp-1YI: |
| Source |
|
 Keywords Keywords | PROTEIN TRANSPORT / glucose transporter / SGLT / sodium glucose transporter / membrane protein |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers





 homo sapiens (human)
homo sapiens (human)