+Search query
-Structure paper
| Title | Structure, gating, and pharmacology of human Ca3.3 channel. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 13, Issue 1, Page 2084, Year 2022 |
| Publish date | Apr 19, 2022 |
 Authors Authors | Lingli He / Zhuoya Yu / Ze Geng / Zhuo Huang / Changjiang Zhang / Yanli Dong / Yiwei Gao / Yuhang Wang / Qihao Chen / Le Sun / Xinyue Ma / Bo Huang / Xiaoqun Wang / Yan Zhao /  |
| PubMed Abstract | The low-voltage activated T-type calcium channels regulate cellular excitability and oscillatory behavior of resting membrane potential which trigger many physiological events and have been ...The low-voltage activated T-type calcium channels regulate cellular excitability and oscillatory behavior of resting membrane potential which trigger many physiological events and have been implicated with many diseases. Here, we determine structures of the human T-type Ca3.3 channel, in the absence and presence of antihypertensive drug mibefradil, antispasmodic drug otilonium bromide and antipsychotic drug pimozide. Ca3.3 contains a long bended S6 helix from domain III, with a positive charged region protruding into the cytosol, which is critical for T-type Ca channel activation at low voltage. The drug-bound structures clearly illustrate how these structurally different compounds bind to the same central cavity inside the Ca3.3 channel, but are mediated by significantly distinct interactions between drugs and their surrounding residues. Phospholipid molecules penetrate into the central cavity in various extent to shape the binding pocket and play important roles in stabilizing the inhibitor. These structures elucidate mechanisms of channel gating, drug recognition, and actions, thus pointing the way to developing potent and subtype-specific drug for therapeutic treatments of related disorders. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:35440630 / PubMed:35440630 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.3 - 3.9 Å |
| Structure data | EMDB-32584, PDB-7wli: EMDB-32585, PDB-7wlj: EMDB-32586, PDB-7wlk: EMDB-32587, PDB-7wll: |
| Chemicals |  ChemComp-CA:  ChemComp-NAG:  ChemComp-3PE:  ChemComp-Y01:  ChemComp-MWV: 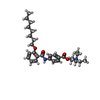 ChemComp-7TB: 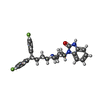 ChemComp-1II: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / apo state / MIB / OB / CaV3.3 / PMZ-bound |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers











 homo sapiens (human)
homo sapiens (human)