+Search query
-Structure paper
| Title | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants. |
|---|---|
| Journal, issue, pages | Nature, Vol. 603, Issue 7903, Page 919-925, Year 2022 |
| Publish date | Jan 28, 2022 |
 Authors Authors | Kang Wang / Zijing Jia / Linilin Bao / Lei Wang / Lei Cao / Hang Chi / Yaling Hu / Qianqian Li / Yunjiao Zhou / Yinan Jiang / Qianhui Zhu / Yongqiang Deng / Pan Liu / Nan Wang / Lin Wang / Min Liu / Yurong Li / Boling Zhu / Kaiyue Fan / Wangjun Fu / Peng Yang / Xinran Pei / Zhen Cui / Lili Qin / Pingju Ge / Jiajing Wu / Shuo Liu / Yiding Chen / Weijin Huang / Qiao Wang / Cheng-Feng Qin / Youchun Wang / Chuan Qin / Xiangxi Wang /  |
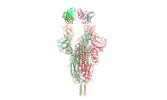
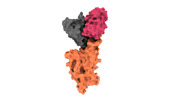
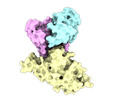
| PubMed Abstract | Omicron (B.1.1.529), the most heavily mutated SARS-CoV-2 variant so far, is highly resistant to neutralizing antibodies, raising concerns about the effectiveness of antibody therapies and vaccines. ...Omicron (B.1.1.529), the most heavily mutated SARS-CoV-2 variant so far, is highly resistant to neutralizing antibodies, raising concerns about the effectiveness of antibody therapies and vaccines. Here we examined whether sera from individuals who received two or three doses of inactivated SARS-CoV-2 vaccine could neutralize authentic Omicron. The seroconversion rates of neutralizing antibodies were 3.3% (2 out of 60) and 95% (57 out of 60) for individuals who had received 2 and 3 doses of vaccine, respectively. For recipients of three vaccine doses, the geometric mean neutralization antibody titre for Omicron was 16.5-fold lower than for the ancestral virus (254). We isolated 323 human monoclonal antibodies derived from memory B cells in triple vaccinees, half of which recognized the receptor-binding domain, and showed that a subset (24 out of 163) potently neutralized all SARS-CoV-2 variants of concern, including Omicron. Therapeutic treatments with representative broadly neutralizing monoclonal antibodies were highly protective against infection of mice with SARS-CoV-2 Beta (B.1.351) and Omicron. Atomic structures of the Omicron spike protein in complex with three classes of antibodies that were active against all five variants of concern defined the binding and neutralizing determinants and revealed a key antibody escape site, G446S, that confers greater resistance to a class of antibodies that bind on the right shoulder of the receptor-binding domain by altering local conformation at the binding interface. Our results rationalize the use of three-dose immunization regimens and suggest that the fundamental epitopes revealed by these broadly ultrapotent antibodies are rational targets for a universal sarbecovirus vaccine. |
 External links External links |  Nature / Nature /  PubMed:35090164 / PubMed:35090164 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.3 - 4.0 Å |
| Structure data | EMDB-32441, PDB-7we7: EMDB-32442, PDB-7we8: EMDB-32443, PDB-7we9: EMDB-32444, PDB-7wea: EMDB-32445, PDB-7web: EMDB-32446, PDB-7wec: EMDB-32447, PDB-7wed: EMDB-32448, PDB-7wee: EMDB-32449, PDB-7wef: EMDB-32581, PDB-7wlc: |
| Chemicals |  ChemComp-NAG: |
| Source |
|
 Keywords Keywords | IMMUNE SYSTEM/VIRAL PROTEIN / SARS-CoV-2 / Omicron / Spike-Fab complex / VIRAL PROTEIN / IMMUNE SYSTEM-VIRAL PROTEIN complex / VIRAL PROTEIN/IMMUNE SYSTEM / VIRAL PROTEIN-IMMUNE SYSTEM complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




















 homo sapiens (human)
homo sapiens (human)
