+Search query
-Structure paper
| Title | Structural Basis of the Modulation of the Voltage-Gated Calcium Ion Channel Ca 1.1 by Dihydropyridine Compounds*. |
|---|---|
| Journal, issue, pages | Angew Chem Int Ed Engl, Vol. 60, Issue 6, Page 3131-3137, Year 2021 |
| Publish date | Feb 8, 2021 |
 Authors Authors | Shuai Gao / Nieng Yan /  |
| PubMed Abstract | 1,4-Dihydropyridines (DHP), the most commonly used antihypertensives, function by inhibiting the L-type voltage-gated Ca (Ca ) channels. DHP compounds exhibit chirality-specific antagonistic or ...1,4-Dihydropyridines (DHP), the most commonly used antihypertensives, function by inhibiting the L-type voltage-gated Ca (Ca ) channels. DHP compounds exhibit chirality-specific antagonistic or agonistic effects. The structure of rabbit Ca 1.1 bound to an achiral drug nifedipine reveals the general binding mode for DHP drugs, but the molecular basis for chiral specificity remained elusive. Herein, we report five cryo-EM structures of nanodisc-embedded Ca 1.1 in the presence of the bestselling drug amlodipine, a DHP antagonist (R)-(+)-Bay K8644, and a titration of its agonistic enantiomer (S)-(-)-Bay K8644 at resolutions of 2.9-3.4 Å. The amlodipine-bound structure reveals the molecular basis for the high efficacy of the drug. All structures with the addition of the Bay K8644 enantiomers exhibit similar inactivated conformations, suggesting that (S)-(-)-Bay K8644, when acting as an agonist, is insufficient to lock the activated state of the channel for a prolonged duration. |
 External links External links |  Angew Chem Int Ed Engl / Angew Chem Int Ed Engl /  PubMed:33125829 / PubMed:33125829 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.9 - 3.4 Å |
| Structure data | EMDB-22414, PDB-7jpk: EMDB-22415, PDB-7jpl: EMDB-22424, PDB-7jpv: EMDB-22425, PDB-7jpw: EMDB-22426, PDB-7jpx: |
| Chemicals |  ChemComp-CA:  ChemComp-3PE:  ChemComp-PC1: 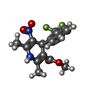 ChemComp-C8U: 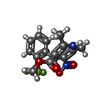 ChemComp-VFY:  ChemComp-POV: 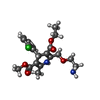 ChemComp-6UB: |
| Source |
|
 Keywords Keywords | TRANSPORT PROTEIN / rCav1.1 / Channels; Calcium Ion-Selective / drugs / Channels / Calcium Ion-Selective |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers














