+Search query
-Structure paper
| Title | Cryo-EM Structures of Chronic Traumatic Encephalopathy Tau Filaments with PET Ligand Flortaucipir. |
|---|---|
| Journal, issue, pages | J Mol Biol, Vol. 435, Issue 11, Page 168025, Year 2023 |
| Publish date | Jun 1, 2023 |
 Authors Authors | Yang Shi / Bernardino Ghetti / Michel Goedert / Sjors H W Scheres /   |
| PubMed Abstract | Positron emission tomography (PET) imaging allows monitoring the progression of amyloid aggregation in the living brain. [F]-Flortaucipir is the only approved PET tracer compound for the ...Positron emission tomography (PET) imaging allows monitoring the progression of amyloid aggregation in the living brain. [F]-Flortaucipir is the only approved PET tracer compound for the visualisation of tau aggregation. Here, we describe cryo-EM experiments on tau filaments in the presence and absence of flortaucipir. We used tau filaments isolated from the brain of an individual with Alzheimer's disease (AD), and from the brain of an individual with primary age-related tauopathy (PART) with a co-pathology of chronic traumatic encephalopathy (CTE). Unexpectedly, we were unable to visualise additional cryo-EM density for flortaucipir for AD paired helical or straight filaments (PHFs or SFs), but we did observe density for flortaucipir binding to CTE Type I filaments from the case with PART. In the latter, flortaucipir binds in a 1:1 molecular stoichiometry with tau, adjacent to lysine 353 and aspartate 358. By adopting a tilted geometry with respect to the helical axis, the 4.7 Å distance between neighbouring tau monomers is reconciled with the 3.5 Å distance consistent with π-π-stacking between neighbouring molecules of flortaucipir. |
 External links External links |  J Mol Biol / J Mol Biol /  PubMed:37330290 / PubMed:37330290 /  PubMed Central PubMed Central |
| Methods | EM (helical sym.) |
| Resolution | 2.6 - 3.66 Å |
| Structure data | EMDB-16329, PDB-8byn:  EMDB-35403: Paired helical filament from Alzheimer's disease brain with PET ligand flortaucipir (+flortaucipir)  EMDB-35404: Paired helical filament from Alzheimer's disease brain incubated with DMSO without PET ligand flortaucipir (-flortaucipir)  EMDB-35405: Paired helical filament from primary age-related tauopathy brain with PET ligand flortaucipir (+flortaucipir)  EMDB-35406: Straight filament from primary age-related tauopathy brain with PET ligand flortaucipir (+flortaucipir)  EMDB-35407: Paired helical filament from primary age-related tauopathy brain incubated with DMSO without PET ligand flortaucipir (-flortaucipir)  EMDB-35408: Straight filament from primary age-related tauopathy brain incubated with DMSO without PET ligand flortaucipir (-flortaucipir)  EMDB-35409: Chronic traumatic encephalopathy type I tau filaments from primary age-related tauopathy brain incubated with DMSO without PET ligand flortaucipir (-flortaucipir) |
| Chemicals | 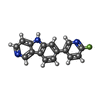 ChemComp-S9C: |
| Source |
|
 Keywords Keywords | PROTEIN FIBRIL / Chronic traumatic encephalopathy / Primary age-related tauopathy / Tau filaments / PET ligand / Flortaucipir |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




 homo sapiens (human)
homo sapiens (human)