+Search query
-Structure paper
| Title | The crystal structure of netrin-1 in complex with DCC reveals the bifunctionality of netrin-1 as a guidance cue. |
|---|---|
| Journal, issue, pages | Neuron, Vol. 83, Issue 4, Page 839-849, Year 2014 |
| Publish date | Aug 20, 2014 |
 Authors Authors | Lorenzo I Finci / Nina Krüger / Xiaqin Sun / Jie Zhang / Magda Chegkazi / Yu Wu / Gundolf Schenk / Haydyn D T Mertens / Dmitri I Svergun / Yan Zhang / Jia-Huai Wang / Rob Meijers /    |
| PubMed Abstract | Netrin-1 is a guidance cue that can trigger either attraction or repulsion effects on migrating axons of neurons, depending on the repertoire of receptors available on the growth cone. How a single ...Netrin-1 is a guidance cue that can trigger either attraction or repulsion effects on migrating axons of neurons, depending on the repertoire of receptors available on the growth cone. How a single chemotropic molecule can act in such contradictory ways has long been a puzzle at the molecular level. Here we present the crystal structure of netrin-1 in complex with the Deleted in Colorectal Cancer (DCC) receptor. We show that one netrin-1 molecule can simultaneously bind to two DCC molecules through a DCC-specific site and through a unique generic receptor binding site, where sulfate ions staple together positively charged patches on both DCC and netrin-1. Furthermore, we demonstrate that UNC5A can replace DCC on the generic receptor binding site to switch the response from attraction to repulsion. We propose that the modularity of binding allows for the association of other netrin receptors at the generic binding site, eliciting alternative turning responses. |
 External links External links |  Neuron / Neuron /  PubMed:25123307 / PubMed:25123307 /  PubMed Central PubMed Central |
| Methods | SAS (X-ray synchrotron) / X-ray diffraction |
| Resolution | 3.1 Å |
| Structure data |  SASDA26:  SASDA76: NetrinVIV DCC56 complex (Netrin-1, NetrinVIV + Deleted in Colorectal Cancer (FN5 & FN6), dcc56) 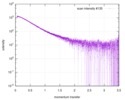 SASDA86: NetrinVIV DCC56(M933R) complex (Netrin-1, NetrinVIV + Deleted in Colorectal Cancer (FN5 & FN6) M933R mutant, DCC56 (M933R))  SASDAZ5:  PDB-4urt: |
| Chemicals |  ChemComp-CA:  ChemComp-CL:  ChemComp-SO4:  ChemComp-GOL: |
| Source |
|
 Keywords Keywords | PROTEIN BINDING / APOPTOSIS / AXON GUIDANCE / DEPENDENCE RECEPTOR |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 homo sapiens (human)
homo sapiens (human)