+Search query
-Structure paper
| Title | Single hormone or synthetic agonist induces G/G coupling selectivity of EP receptors via distinct binding modes and propagating paths. |
|---|---|
| Journal, issue, pages | Proc Natl Acad Sci U S A, Vol. 120, Issue 30, Page e2216329120, Year 2023 |
| Publish date | Jul 25, 2023 |
 Authors Authors | Shen-Ming Huang / Meng-Yao Xiong / Lei Liu / Jianqiang Mu / Ming-Wei Wang / Ying-Li Jia / Kui Cai / Lu Tie / Chao Zhang / Sheng Cao / Xin Wen / Jia-Le Wang / Sheng-Chao Guo / Yu Li / Chang-Xiu Qu / Qing-Tao He / Bo-Yang Cai / Chenyang Xue / Shiyi Gan / Yihe Xie / Xin Cong / Zhao Yang / Wei Kong / Shuo Li / Zijian Li / Peng Xiao / Fan Yang / Xiao Yu / You-Fei Guan / Xiaoyan Zhang / Zhongmin Liu / Bao-Xue Yang / Yang Du / Jin-Peng Sun /  |
| PubMed Abstract | To accomplish concerted physiological reactions, nature has diversified functions of a single hormone at at least two primary levels: 1) Different receptors recognize the same hormone, and 2) ...To accomplish concerted physiological reactions, nature has diversified functions of a single hormone at at least two primary levels: 1) Different receptors recognize the same hormone, and 2) different cellular effectors couple to the same hormone-receptor pair [R.P. Xiao, , re15 (2001); L. Hein, J. D. Altman, B.K. Kobilka, , 181-184 (1999); Y. Daaka, L. M. Luttrell, R. J. Lefkowitz, , 88-91 (1997)]. Not only these questions lie in the heart of hormone actions and receptor signaling but also dissecting mechanisms underlying these questions could offer therapeutic routes for refractory diseases, such as kidney injury (KI) or X-linked nephrogenic diabetes insipidus (NDI). Here, we identified that G-biased signaling, but not G activation downstream of EP4, showed beneficial effects for both KI and NDI treatments. Notably, by solving Cryo-electron microscope (cryo-EM) structures of EP3-G, EP4-G, and EP4-G in complex with endogenous prostaglandin E (PGE)or two synthetic agonists and comparing with PGE-EP2-G structures, we found that unique primary sequences of prostaglandin E2 receptor (EP) receptors and distinct conformational states of the EP4 ligand pocket govern the G/G transducer coupling selectivity through different structural propagation paths, especially via TM6 and TM7, to generate selective cytoplasmic structural features. In particular, the orientation of the PGE ω-chain and two distinct pockets encompassing agonist L902688 of EP4 were differentiated by their G/G coupling ability. Further, we identified common and distinct features of cytoplasmic side of EP receptors for G/G coupling and provide a structural basis for selective and biased agonist design of EP4 with therapeutic potential. |
 External links External links |  Proc Natl Acad Sci U S A / Proc Natl Acad Sci U S A /  PubMed:37478163 / PubMed:37478163 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.1 - 3.5 Å |
| Structure data | EMDB-29935, PDB-8gcm: EMDB-29940, PDB-8gcp: EMDB-29943, PDB-8gd9: EMDB-29944, PDB-8gda: EMDB-29945, PDB-8gdb: EMDB-29946, PDB-8gdc: |
| Chemicals | 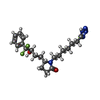 ChemComp-YX9: 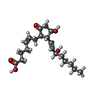 ChemComp-P2E: 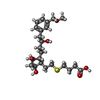 ChemComp-Z2C:  ChemComp-YZV: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / GPCR complex / SIGNALING PROTEIN |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers















 homo sapiens (human)
homo sapiens (human)
