+Search query
-Structure paper
| Title | A surface on the androgen receptor that allosterically regulates coactivator binding. |
|---|---|
| Journal, issue, pages | Proc. Natl. Acad. Sci. Usa, Vol. 104, Page 16074-16079, Year 2007 |
| Publish date | Apr 13, 2007 (structure data deposition date) |
 Authors Authors | Estebanez-Perpina, E. / Arnold, L.A. / Arnold, A.A. / Nguyen, P. / Rodrigues, E.D. / Mar, E. / Bateman, R. / Pallai, P. / Shokat, K.M. / Baxter, J.D. ...Estebanez-Perpina, E. / Arnold, L.A. / Arnold, A.A. / Nguyen, P. / Rodrigues, E.D. / Mar, E. / Bateman, R. / Pallai, P. / Shokat, K.M. / Baxter, J.D. / Guy, R.K. / Webb, P. / Fletterick, R.J. |
 External links External links |  Proc. Natl. Acad. Sci. Usa / Proc. Natl. Acad. Sci. Usa /  PubMed:17911242 PubMed:17911242 |
| Methods | X-ray diffraction |
| Resolution | 1.76 - 2.58 Å |
| Structure data |  PDB-2pio:  PDB-2pip:  PDB-2piq:  PDB-2pir:  PDB-2pit:  PDB-2piu:  PDB-2piv:  PDB-2piw:  PDB-2pix:  PDB-2pkl:  PDB-2qpy: |
| Chemicals |  ChemComp-SO4:  ChemComp-DHT:  ChemComp-2MI:  ChemComp-HOH:  ChemComp-ICO:  ChemComp-K10:  ChemComp-RB1:  ChemComp-NK:  ChemComp-4HY: 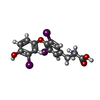 ChemComp-T3: 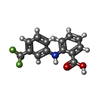 ChemComp-FLF: |
| Source |
|
 Keywords Keywords | HORMONE RECEPTOR / androgen receptor ligand binding domain / inhibitors coactivator binding AF-2 pocket / androgen receptor / coactivators / AF-2 pocket / inhibitors / inhibitors coactivator binding AF-2 / androgen receptor coactivator binding inhibitors AF-2 pocket / androgen receptor coactivator binding inhibitors AF-2 / androgen receptor inhibitors coactivator binding AF-2 / androgen receptor inhibitor coactivator binding / androgen receptor inhibitors coactivator binding / AR LBD ihnibitors coactivator binding / androgen receptor coactivators AF2 inhibitors / DNA BINDING PROTEIN / androgen receptor DHT coactivator / DNA-binding / Lipid-binding / Metal-binding / Nucleus / Steroid-binding / Transcription / Transcription regulation / Ubl conjugation / Zinc / Zinc-finger |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 homo sapiens (human)
homo sapiens (human)
