+Search query
-Structure paper
| Title | Structural insights into opposing actions of neurosteroids on GABA receptors. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 14, Issue 1, Page 5091, Year 2023 |
| Publish date | Aug 22, 2023 |
 Authors Authors | Dagimhiwat H Legesse / Chen Fan / Jinfeng Teng / Yuxuan Zhuang / Rebecca J Howard / Colleen M Noviello / Erik Lindahl / Ryan E Hibbs /   |
| PubMed Abstract | γ-Aminobutyric acid type A (GABA) receptors mediate fast inhibitory signaling in the brain and are targets of numerous drugs and endogenous neurosteroids. A subset of neurosteroids are GABA receptor ...γ-Aminobutyric acid type A (GABA) receptors mediate fast inhibitory signaling in the brain and are targets of numerous drugs and endogenous neurosteroids. A subset of neurosteroids are GABA receptor positive allosteric modulators; one of these, allopregnanolone, is the only drug approved specifically for treating postpartum depression. There is a consensus emerging from structural, physiological and photolabeling studies as to where positive modulators bind, but how they potentiate GABA activation remains unclear. Other neurosteroids are negative modulators of GABA receptors, but their binding sites remain debated. Here we present structures of a synaptic GABA receptor bound to allopregnanolone and two inhibitory sulfated neurosteroids. Allopregnanolone binds at the receptor-bilayer interface, in the consensus potentiator site. In contrast, inhibitory neurosteroids bind in the pore. MD simulations and electrophysiology support a mechanism by which allopregnanolone potentiates channel activity and suggest the dominant mechanism for sulfated neurosteroid inhibition is through pore block. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:37607940 / PubMed:37607940 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.65 - 2.98 Å |
| Structure data | EMDB-40462, PDB-8sgo: EMDB-40503, PDB-8si9: EMDB-40506, PDB-8sid: |
| Chemicals |  ChemComp-NAG:  ChemComp-ABU:  ChemComp-PTY:  ChemComp-Q3G:  ChemComp-A8W: 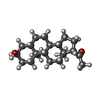 ChemComp-Y4B: 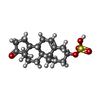 ChemComp-ZWY:  ChemComp-POV: |
| Source |
|
 Keywords Keywords | TRANSPORT PROTEIN/IMMUNE SYSTEM / Ion channel / neurosteroids / TRANSPORT PROTEIN-IMMUNE SYSTEM complex / TRANSPORT PROTEIN |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers









 homo sapiens (human)
homo sapiens (human)
