+Search query
-Structure paper
| Title | Recognition of cyclic dinucleotides and folates by human SLC19A1. |
|---|---|
| Journal, issue, pages | Nature, Vol. 612, Issue 7938, Page 170-176, Year 2022 |
| Publish date | Oct 20, 2022 |
 Authors Authors | Qixiang Zhang / Xuyuan Zhang / Yalan Zhu / Panpan Sun / Liwei Zhang / Junxiao Ma / Yong Zhang / Lingan Zeng / Xiaohua Nie / Yina Gao / Zhaolong Li / Songqing Liu / Jizhong Lou / Ang Gao / Liguo Zhang / Pu Gao /  |
| PubMed Abstract | Cyclic dinucleotides (CDNs) are ubiquitous signalling molecules in all domains of life. Mammalian cells produce one CDN, 2'3'-cGAMP, through cyclic GMP-AMP synthase after detecting cytosolic DNA ...Cyclic dinucleotides (CDNs) are ubiquitous signalling molecules in all domains of life. Mammalian cells produce one CDN, 2'3'-cGAMP, through cyclic GMP-AMP synthase after detecting cytosolic DNA signals. 2'3'-cGAMP, as well as bacterial and synthetic CDN analogues, can act as second messengers to activate stimulator of interferon genes (STING) and elicit broad downstream responses. Extracellular CDNs must traverse the cell membrane to activate STING, a process that is dependent on the solute carrier SLC19A1. Moreover, SLC19A1 represents the major transporter for folate nutrients and antifolate therapeutics, thereby placing SLC19A1 as a key factor in multiple physiological and pathological processes. How SLC19A1 recognizes and transports CDNs, folate and antifolate is unclear. Here we report cryo-electron microscopy structures of human SLC19A1 (hSLC19A1) in a substrate-free state and in complexes with multiple CDNs from different sources, a predominant natural folate and a new-generation antifolate drug. The structural and mutagenesis results demonstrate that hSLC19A1 uses unique yet divergent mechanisms to recognize CDN- and folate-type substrates. Two CDN molecules bind within the hSLC19A1 cavity as a compact dual-molecule unit, whereas folate and antifolate bind as a monomer and occupy a distinct pocket of the cavity. Moreover, the structures enable accurate mapping and potential mechanistic interpretation of hSLC19A1 with loss-of-activity and disease-related mutations. Our research provides a framework for understanding the mechanism of SLC19-family transporters and is a foundation for the development of potential therapeutics. |
 External links External links |  Nature / Nature /  PubMed:36265513 PubMed:36265513 |
| Methods | EM (single particle) |
| Resolution | 3.0 - 3.4 Å |
| Structure data | EMDB-33386, PDB-7xpz: EMDB-33387, PDB-7xq0: EMDB-33388, PDB-7xq1: EMDB-33389, PDB-7xq2: EMDB-34176, PDB-8goe: EMDB-34177, PDB-8gof: |
| Chemicals | 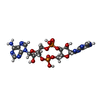 ChemComp-2BA: 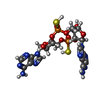 ChemComp-GJF: 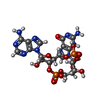 ChemComp-1SY:  ChemComp-THH:  ChemComp-LYA: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / SLC19A1 / hSLC19A1+3'3'-CDA complex / SLC19A1+2'3'-CDAS complex / hSLC19A1+2'3'-cGAMP complex / hSLC19A1+5-MTHF complex / hSLC19A1+PMX complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers















 homo sapiens (human)
homo sapiens (human)