+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
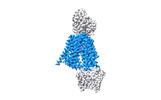
| Title | Structure of hSLC19A1+5-MTHF | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | hSLC19A1+5-MTHF complex / MEMBRANE PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationfolate:monoatomic anion antiporter activity / methotrexate transport / folic acid transmembrane transporter activity / methotrexate transmembrane transporter activity / folic acid transport / folate transmembrane transport / folate import across plasma membrane / cyclic-GMP-AMP transmembrane transporter activity / cyclic-GMP-AMP transmembrane import across plasma membrane / Metabolism of folate and pterines ...folate:monoatomic anion antiporter activity / methotrexate transport / folic acid transmembrane transporter activity / methotrexate transmembrane transporter activity / folic acid transport / folate transmembrane transport / folate import across plasma membrane / cyclic-GMP-AMP transmembrane transporter activity / cyclic-GMP-AMP transmembrane import across plasma membrane / Metabolism of folate and pterines / 2',3'-cyclic GMP-AMP binding / positive regulation of cGAS/STING signaling pathway / xenobiotic transmembrane transport / organic anion transport / : / folic acid binding / antiporter activity / folic acid metabolic process / xenobiotic transmembrane transporter activity / transport across blood-brain barrier / brush border membrane / female pregnancy / response to toxic substance / basolateral plasma membrane / apical plasma membrane / response to xenobiotic stimulus / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | ||||||||||||||||||
 Authors Authors | Zhang QX / Zhang XY / Zhu YL / Sun PP / Gao A / Zhang LG / Gao P | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Recognition of cyclic dinucleotides and folates by human SLC19A1. Authors: Qixiang Zhang / Xuyuan Zhang / Yalan Zhu / Panpan Sun / Liwei Zhang / Junxiao Ma / Yong Zhang / Lingan Zeng / Xiaohua Nie / Yina Gao / Zhaolong Li / Songqing Liu / Jizhong Lou / Ang Gao / Liguo Zhang / Pu Gao /  Abstract: Cyclic dinucleotides (CDNs) are ubiquitous signalling molecules in all domains of life. Mammalian cells produce one CDN, 2'3'-cGAMP, through cyclic GMP-AMP synthase after detecting cytosolic DNA ...Cyclic dinucleotides (CDNs) are ubiquitous signalling molecules in all domains of life. Mammalian cells produce one CDN, 2'3'-cGAMP, through cyclic GMP-AMP synthase after detecting cytosolic DNA signals. 2'3'-cGAMP, as well as bacterial and synthetic CDN analogues, can act as second messengers to activate stimulator of interferon genes (STING) and elicit broad downstream responses. Extracellular CDNs must traverse the cell membrane to activate STING, a process that is dependent on the solute carrier SLC19A1. Moreover, SLC19A1 represents the major transporter for folate nutrients and antifolate therapeutics, thereby placing SLC19A1 as a key factor in multiple physiological and pathological processes. How SLC19A1 recognizes and transports CDNs, folate and antifolate is unclear. Here we report cryo-electron microscopy structures of human SLC19A1 (hSLC19A1) in a substrate-free state and in complexes with multiple CDNs from different sources, a predominant natural folate and a new-generation antifolate drug. The structural and mutagenesis results demonstrate that hSLC19A1 uses unique yet divergent mechanisms to recognize CDN- and folate-type substrates. Two CDN molecules bind within the hSLC19A1 cavity as a compact dual-molecule unit, whereas folate and antifolate bind as a monomer and occupy a distinct pocket of the cavity. Moreover, the structures enable accurate mapping and potential mechanistic interpretation of hSLC19A1 with loss-of-activity and disease-related mutations. Our research provides a framework for understanding the mechanism of SLC19-family transporters and is a foundation for the development of potential therapeutics. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34176.map.gz emd_34176.map.gz | 117.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34176-v30.xml emd-34176-v30.xml emd-34176.xml emd-34176.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34176.png emd_34176.png | 36.2 KB | ||
| Filedesc metadata |  emd-34176.cif.gz emd-34176.cif.gz | 5.4 KB | ||
| Others |  emd_34176_half_map_1.map.gz emd_34176_half_map_1.map.gz emd_34176_half_map_2.map.gz emd_34176_half_map_2.map.gz | 98.5 MB 98.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34176 http://ftp.pdbj.org/pub/emdb/structures/EMD-34176 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34176 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34176 | HTTPS FTP |
-Related structure data
| Related structure data |  8goeMC  7xpzC  7xq0C  7xq1C  7xq2C  8gofC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34176.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34176.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34176_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_34176_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SLC19A1+5-MTHF
| Entire | Name: SLC19A1+5-MTHF |
|---|---|
| Components |
|
-Supramolecule #1: SLC19A1+5-MTHF
| Supramolecule | Name: SLC19A1+5-MTHF / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Reduced folate transporter
| Macromolecule | Name: Reduced folate transporter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 60.556082 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSMVPSSPA VEKQVPVEPG PDPELRSWRH LVCYLCFYGF MAQIRPGESF ITPYLLGPDK NFTREQVTNE ITPVLSYSYL AVLVPVFLL TDYLRYTPVL LLQGLSFVSV WLLLLLGHSV AHMQLMELFY SVTMAARIAY SSYIFSLVRP ARYQRVAGYS R AAVLLGVF ...String: MGSMVPSSPA VEKQVPVEPG PDPELRSWRH LVCYLCFYGF MAQIRPGESF ITPYLLGPDK NFTREQVTNE ITPVLSYSYL AVLVPVFLL TDYLRYTPVL LLQGLSFVSV WLLLLLGHSV AHMQLMELFY SVTMAARIAY SSYIFSLVRP ARYQRVAGYS R AAVLLGVF TSSVLGQLLV TVGRVSFSTL NYISLAFLTF SVVLALFLKR PKRSLFFNRD DRGRCETSAS ELERMNPGPG GK LGHALRV ACGDSVLARM LRELGDSLRR PQLRLWSLWW VFNSAGYYLV VYYVHILWNE VDPTTNSARV YNGAADAAST LLG AITSFA AGFVKIRWAR WSKLLIAGVT ATQAGLVFLL AHTRHPSSIW LCYAAFVLFR GSYQFLVPIA TFQIASSLSK ELCA LVFGV NTFFATIVKT IITFIVSDVR GLGLPVRKQF QLYSVYFLIL SIIYFLGAML DGLRHCQRGH HPRQPPAQGL RSAAE EKAA QALSVQDKGL GGLQPAQSPP LSPEDKLGSE NLYLEVLFQG PFQGGSGGSG HHHHHHHHHH UniProtKB: Reduced folate transporter |
-Macromolecule #2: N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDI...
| Macromolecule | Name: N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID type: ligand / ID: 2 / Number of copies: 1 / Formula: THH |
|---|---|
| Molecular weight | Theoretical: 459.456 Da |
| Chemical component information |  ChemComp-THH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 182748 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)