+Search query
-Structure paper
| Title | Structures of the human cholecystokinin receptors bound to agonists and antagonists. |
|---|---|
| Journal, issue, pages | Nat Chem Biol, Vol. 17, Issue 12, Page 1230-1237, Year 2021 |
| Publish date | Sep 23, 2021 |
 Authors Authors | Xuefeng Zhang / Chenglin He / Mu Wang / Qingtong Zhou / Dehua Yang / Ya Zhu / Wenbo Feng / Hui Zhang / Antao Dai / Xiaojing Chu / Jia Wang / Zhenlin Yang / Yi Jiang / Ulrich Sensfuss / Qiuxiang Tan / Shuo Han / Steffen Reedtz-Runge / H Eric Xu / Suwen Zhao / Ming-Wei Wang / Beili Wu / Qiang Zhao /   |
| PubMed Abstract | Cholecystokinin receptors, CCKR and CCKR, are important neurointestinal peptide hormone receptors and play a vital role in food intake and appetite regulation. Here, we report three crystal ...Cholecystokinin receptors, CCKR and CCKR, are important neurointestinal peptide hormone receptors and play a vital role in food intake and appetite regulation. Here, we report three crystal structures of the human CCKR in complex with different ligands, including one peptide agonist and two small-molecule antagonists, as well as two cryo-electron microscopy structures of CCKR-gastrin in complex with G and G, respectively. These structures reveal the recognition pattern of different ligand types and the molecular basis of peptide selectivity in the cholecystokinin receptor family. By comparing receptor structures in different conformational states, a stepwise activation process of cholecystokinin receptors is proposed. Combined with pharmacological data, our results provide atomic details for differential ligand recognition and receptor activation mechanisms. These insights will facilitate the discovery of potential therapeutics targeting cholecystokinin receptors. |
 External links External links |  Nat Chem Biol / Nat Chem Biol /  PubMed:34556863 PubMed:34556863 |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 2.5 - 3.3 Å |
| Structure data | EMDB-31493, PDB-7f8v: EMDB-31494, PDB-7f8w:  PDB-7f8u:  PDB-7f8x:  PDB-7f8y: |
| Chemicals | 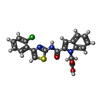 ChemComp-1OE: 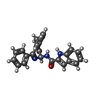 ChemComp-1OZ: |
| Source |
|
 Keywords Keywords | STRUCTURAL PROTEIN / G protein-coulped receptor / Cholecystokinin receptor CCKAR / lintitript / Cholecystokinin receptor CCKBR / Gastrin-17 / devazepide |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers







 homo sapiens (human)
homo sapiens (human) enterobacteria phage t4 (virus)
enterobacteria phage t4 (virus)