+Search query
-Structure paper
| Title | Mechanism of substrate transport and inhibition of the human LAT1-4F2hc amino acid transporter. |
|---|---|
| Journal, issue, pages | Cell Discov, Vol. 7, Issue 1, Page 16, Year 2021 |
| Publish date | Mar 23, 2021 |
 Authors Authors | Renhong Yan / Yaning Li / Jennifer Müller / Yuanyuan Zhang / Simon Singer / Lu Xia / Xinyue Zhong / Jürg Gertsch / Karl-Heinz Altmann / Qiang Zhou /   |
| PubMed Abstract | LAT1 (SLC7A5) is one of the representative light chain proteins of heteromeric amino acid transporters, forming a heterodimer with its heavy chain partner 4F2hc (SLC3A2). LAT1 is overexpressed in ...LAT1 (SLC7A5) is one of the representative light chain proteins of heteromeric amino acid transporters, forming a heterodimer with its heavy chain partner 4F2hc (SLC3A2). LAT1 is overexpressed in many types of tumors and mediates the transfer of drugs and hormones across the blood-brain barrier. Thus, LAT1 is considered as a drug target for cancer treatment and may be exploited for drug delivery into the brain. Here, we synthesized three potent inhibitors of human LAT1, which inhibit transport of leucine with IC values between 100 and 250 nM, and solved the cryo-EM structures of the corresponding LAT1-4F2hc complexes with these inhibitors bound at resolution of up to 2.7 or 2.8 Å. The protein assumes an outward-facing occluded conformation, with the inhibitors bound in the classical substrate binding pocket, but with their tails wedged between the substrate binding site and TM10 of LAT1. We also solved the complex structure of LAT1-4F2hc with 3,5-diiodo-L-tyrosine (Diiodo-Tyr) at 3.4 Å overall resolution, which revealed a different inhibition mechanism and might represent an intermediate conformation between the outward-facing occluded state mentioned above and the outward-open state. To our knowledge, this is the first time that the outward-facing conformation is revealed for the HAT family. Our results unveil more important insights into the working mechanisms of HATs and provide a structural basis for future drug design. |
 External links External links |  Cell Discov / Cell Discov /  PubMed:33758168 / PubMed:33758168 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.7 - 3.4 Å |
| Structure data | EMDB-30835: cryo EM map of the LAT1-4F2hc bound with JX-075  EMDB-30836: EMDB-30837: cryo EM map of the LAT1-4F2hc bound with JX-078  EMDB-30838: EMDB-30839: cryo EM map of the LAT1-4F2hc bound with JX-119  EMDB-30840: EMDB-30841: cryo EM map of the LAT1-4F2hc bound with 3,5-diiodo-L-tyrosine  EMDB-30842: |
| Chemicals |  ChemComp-3PH: 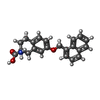 ChemComp-HNX:  ChemComp-Y01:  ChemComp-HOH: 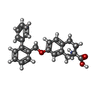 ChemComp-HO0: 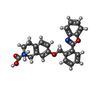 ChemComp-HO9: 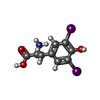 ChemComp-TYI: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / LAT1-4F2hc |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers











 homo sapiens (human)
homo sapiens (human)