+Search query
-Structure paper
| Title | Structural insights into the human D1 and D2 dopamine receptor signaling complexes. |
|---|---|
| Journal, issue, pages | Cell, Vol. 184, Issue 4, Page 931-942.e18, Year 2021 |
| Publish date | Feb 18, 2021 |
 Authors Authors | Youwen Zhuang / Peiyu Xu / Chunyou Mao / Lei Wang / Brian Krumm / X Edward Zhou / Sijie Huang / Heng Liu / Xi Cheng / Xi-Ping Huang / Dan-Dan Shen / Tinghai Xu / Yong-Feng Liu / Yue Wang / Jia Guo / Yi Jiang / Hualiang Jiang / Karsten Melcher / Bryan L Roth / Yan Zhang / Cheng Zhang / H Eric Xu /   |
| PubMed Abstract | The D1- and D2-dopamine receptors (D1R and D2R), which signal through G and G, respectively, represent the principal stimulatory and inhibitory dopamine receptors in the central nervous system. D1R ...The D1- and D2-dopamine receptors (D1R and D2R), which signal through G and G, respectively, represent the principal stimulatory and inhibitory dopamine receptors in the central nervous system. D1R and D2R also represent the main therapeutic targets for Parkinson's disease, schizophrenia, and many other neuropsychiatric disorders, and insight into their signaling is essential for understanding both therapeutic and side effects of dopaminergic drugs. Here, we report four cryoelectron microscopy (cryo-EM) structures of D1R-G and D2R-G signaling complexes with selective and non-selective dopamine agonists, including two currently used anti-Parkinson's disease drugs, apomorphine and bromocriptine. These structures, together with mutagenesis studies, reveal the conserved binding mode of dopamine agonists, the unique pocket topology underlying ligand selectivity, the conformational changes in receptor activation, and potential structural determinants for G protein-coupling selectivity. These results provide both a molecular understanding of dopamine signaling and multiple structural templates for drug design targeting the dopaminergic system. |
 External links External links |  Cell / Cell /  PubMed:33571431 / PubMed:33571431 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.8 - 3.0 Å |
| Structure data | EMDB-22493, PDB-7jv5: EMDB-22509, PDB-7jvp: EMDB-22510, PDB-7jvq: EMDB-22511, PDB-7jvr: |
| Chemicals | 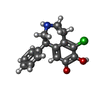 ChemComp-SK0:  ChemComp-CLR:  ChemComp-PLM: 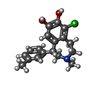 ChemComp-SK9: 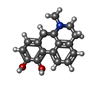 ChemComp-OR9: 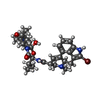 ChemComp-08Y: |
| Source |
|
 Keywords Keywords | SIGNALING PROTEIN / Dopamine receptor 2 / Gi protein / SKF-81297 / Dopamine receptor 1 / SKF-83959 / apomorphine / bromocriptine |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers











 homo sapiens (human)
homo sapiens (human)
