+Search query
-Structure paper
| Title | Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements. |
|---|---|
| Journal, issue, pages | Proc Natl Acad Sci U S A, Vol. 114, Issue 31, Page E6342-E6351, Year 2017 |
| Publish date | Aug 1, 2017 |
 Authors Authors | Gustavo Fuertes / Niccolò Banterle / Kiersten M Ruff / Aritra Chowdhury / Davide Mercadante / Christine Koehler / Michael Kachala / Gemma Estrada Girona / Sigrid Milles / Ankur Mishra / Patrick R Onck / Frauke Gräter / Santiago Esteban-Martín / Rohit V Pappu / Dmitri I Svergun / Edward A Lemke /     |
| PubMed Abstract | Unfolded states of proteins and native states of intrinsically disordered proteins (IDPs) populate heterogeneous conformational ensembles in solution. The average sizes of these heterogeneous ...Unfolded states of proteins and native states of intrinsically disordered proteins (IDPs) populate heterogeneous conformational ensembles in solution. The average sizes of these heterogeneous systems, quantified by the radius of gyration ( ), can be measured by small-angle X-ray scattering (SAXS). Another parameter, the mean dye-to-dye distance ( ) for proteins with fluorescently labeled termini, can be estimated using single-molecule Förster resonance energy transfer (smFRET). A number of studies have reported inconsistencies in inferences drawn from the two sets of measurements for the dimensions of unfolded proteins and IDPs in the absence of chemical denaturants. These differences are typically attributed to the influence of fluorescent labels used in smFRET and to the impact of high concentrations and averaging features of SAXS. By measuring the dimensions of a collection of labeled and unlabeled polypeptides using smFRET and SAXS, we directly assessed the contributions of dyes to the experimental values and For chemically denatured proteins we obtain mutual consistency in our inferences based on and , whereas for IDPs under native conditions, we find substantial deviations. Using computations, we show that discrepant inferences are neither due to methodological shortcomings of specific measurements nor due to artifacts of dyes. Instead, our analysis suggests that chemical heterogeneity in heteropolymeric systems leads to a decoupling between and that is amplified in the absence of denaturants. Therefore, joint assessments of and combined with measurements of polymer shapes should provide a consistent and complete picture of the underlying ensembles. |
 External links External links |  Proc Natl Acad Sci U S A / Proc Natl Acad Sci U S A /  PubMed:28716919 / PubMed:28716919 /  PubMed Central PubMed Central |
| Methods | SAS (X-ray synchrotron) |
| Structure data | 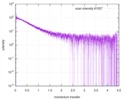 SASDE23: Labeled nuclear pore complex protein Nup153 (NUL-Alexa488/Alexa594) without denaturant  SASDE33: 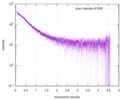 SASDE43: Labeled nuclear pore complex protein Nup153 (NUL-Alexa488/Alexa594) with denaturant 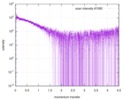 SASDE53: Unlabeled dihydrolipoyllysine-residue succinyltransferase component (BBL) with denaturant 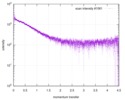 SASDE63: Labeled dihydrolipoyllysine-residue succinyltransferase component (BBL-Alexa488/Alexa594) with denaturant 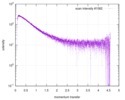 SASDE73: Unlabeled cold shock protein (CSP) with denaturant 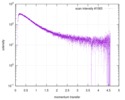 SASDE83: Labeled cold shock protein (CSP-Alexa488/Alexa594) with denaturant 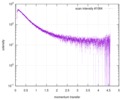 SASDE93: Unlabeled thioredoxin (TRX) with denaturant (Thioredoxin 1, TRX) 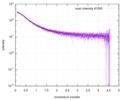 SASDEA3: Labeled thioredoxin (TRX-Alexa488/Alexa594) with denaturant 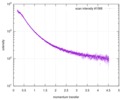 SASDEB3: Unlabeled nuclear pore complex protein Nup98-Nup96 (N98) without denaturant 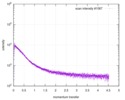 SASDEC3: Unlabeled nucleoporin NSP1 (NSP) without denaturant  SASDEH2: 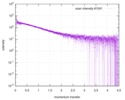 SASDEJ2: Labeled nucleoporin NUP49/NSP49 (N49-Alexa488/Alexa594) without denaturant  SASDEK2: 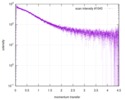 SASDEL2: Labeled nucleoporin NUP49/NSP49 (N49-Alexa488/Alexa594) with denaturant  SASDEM2:  SASDEN2: Labeled Nuclear Localization Signal from the inner nuclear membrane protein HEH2 (NLS-Alexa488/Alexa594) without denaturant  SASDEP2: 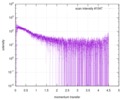 SASDEQ2: Labeled Nuclear Localization Signal from the inner nuclear membrane protein HEH2 (NLS-Alexa488/Alexa594) with denaturant  SASDER2: 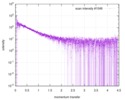 SASDES2: Labeled Importin Beta Binding Domain (IBB-Alexa488/Alexa594) from importin subunit alpha-1 without denaturant  SASDET2:  SASDEU2: Labeled Importin Beta Binding Domain from importin subunit alpha-1 (IBB-Alexa488/Alexa594) with denaturant  SASDEV2: 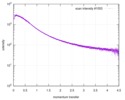 SASDEW2: Labeled nuclear pore complex protein Nup153 (NUS-Alexa488/Alexa594) without denaturant  SASDEX2: 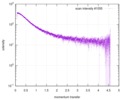 SASDEY2: Labeled nuclear pore complex protein Nup153 (NUS-Alexa488/Alexa594) with denaturant  SASDEZ2: |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 Homo sapiens (human)
Homo sapiens (human)
 Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099) (bacteria)
Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099) (bacteria)
