+Search query
-Structure paper
| Title | Control of transcription elongation and DNA repair by alarmone ppGpp. |
|---|---|
| Journal, issue, pages | Nat Struct Mol Biol, Vol. 30, Issue 5, Page 600-607, Year 2023 |
| Publish date | Mar 30, 2023 |
 Authors Authors | Jacob W Weaver / Sergey Proshkin / Wenqian Duan / Vitaly Epshtein / Manjunath Gowder / Binod K Bharati / Elena Afanaseva / Alexander Mironov / Alexander Serganov / Evgeny Nudler /   |
| PubMed Abstract | Second messenger (p)ppGpp (collectively guanosine tetraphosphate and guanosine pentaphosphate) mediates bacterial adaptation to nutritional stress by modulating transcription initiation. More ...Second messenger (p)ppGpp (collectively guanosine tetraphosphate and guanosine pentaphosphate) mediates bacterial adaptation to nutritional stress by modulating transcription initiation. More recently, ppGpp has been implicated in coupling transcription and DNA repair; however, the mechanism of ppGpp engagement remained elusive. Here we present structural, biochemical and genetic evidence that ppGpp controls Escherichia coli RNA polymerase (RNAP) during elongation via a specific site that is nonfunctional during initiation. Structure-guided mutagenesis renders the elongation (but not initiation) complex unresponsive to ppGpp and increases bacterial sensitivity to genotoxic agents and ultraviolet radiation. Thus, ppGpp binds RNAP at sites with distinct functions in initiation and elongation, with the latter being important for promoting DNA repair. Our data provide insights on the molecular mechanism of ppGpp-mediated adaptation during stress, and further highlight the intricate relationships between genome stability, stress responses and transcription. |
 External links External links |  Nat Struct Mol Biol / Nat Struct Mol Biol /  PubMed:36997761 / PubMed:36997761 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.1 - 2.42 Å |
| Structure data | EMDB-29491, PDB-8fvr: EMDB-29494, PDB-8fvw: |
| Chemicals |  ChemComp-ZN:  ChemComp-MG:  ChemComp-HOH: 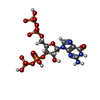 ChemComp-G4P:  ChemComp-NA: |
| Source |
|
 Keywords Keywords | TRANSCRIPTION/DNA/RNA / Transcription elongation / TRANSCRIPTION-DNA-RNA complex / Second messager / ppGpp / DNA repair |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers








