+Search query
-Structure paper
| Title | Cryo-EM structure of the human adenosine A receptor-G signaling complex. |
|---|---|
| Journal, issue, pages | Sci Adv, Vol. 8, Issue 51, Page eadd3709, Year 2022 |
| Publish date | Dec 23, 2022 |
 Authors Authors | Ying Chen / Jinyi Zhang / Yuan Weng / Yueming Xu / Weiqiang Lu / Wei Liu / Mingyao Liu / Tian Hua / Gaojie Song /   |
| PubMed Abstract | The human adenosine A receptor (AR) is a class A G protein-coupled receptor that is involved in several major physiological and pathological processes throughout the body. AR recognizes its ligands ...The human adenosine A receptor (AR) is a class A G protein-coupled receptor that is involved in several major physiological and pathological processes throughout the body. AR recognizes its ligands adenosine and NECA with relatively low affinity, but the detailed mechanism for its ligand recognition and signaling is still elusive. Here, we present two structures determined by cryo-electron microscopy of AR bound to its agonists NECA and BAY60-6583, each coupled to an engineered G protein. The structures reveal conserved orthosteric binding pockets with subtle differences, whereas the selectivity or specificity can mainly be attributed to regions extended from the orthosteric pocket. We also found that BAY60-6583 occupies a secondary pocket, where residues V250 and N273 were two key determinants for its selectivity against AR. This study offers a better understanding of ligand selectivity for the adenosine receptor family and provides a structural template for further development of AR ligands for related diseases. |
 External links External links |  Sci Adv / Sci Adv /  PubMed:36563137 / PubMed:36563137 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.99 - 3.26 Å |
| Structure data | EMDB-33512, PDB-7xy6: EMDB-33513, PDB-7xy7: |
| Chemicals | 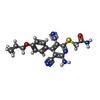 ChemComp-I5D: 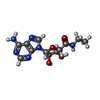 ChemComp-NEC: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / G protein coupled-receptor |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers







 homo sapiens (human)
homo sapiens (human) oplophorus gracilirostris (crustacean)
oplophorus gracilirostris (crustacean)
