[English] 日本語
 Yorodumi
Yorodumi- EMDB-33512: Adenosine receptor bound to an agonist in complex with G protein ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Adenosine receptor bound to an agonist in complex with G protein obtained by cryo-EM | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | G protein coupled-receptor / membrane protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of chronic inflammatory response to non-antigenic stimulus / : / Adenosine P1 receptors / G protein-coupled adenosine receptor activity / positive regulation of mast cell degranulation / Surfactant metabolism / relaxation of vascular associated smooth muscle / : / mast cell degranulation / positive regulation of vascular endothelial growth factor production ...positive regulation of chronic inflammatory response to non-antigenic stimulus / : / Adenosine P1 receptors / G protein-coupled adenosine receptor activity / positive regulation of mast cell degranulation / Surfactant metabolism / relaxation of vascular associated smooth muscle / : / mast cell degranulation / positive regulation of vascular endothelial growth factor production / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / intracellular transport / D1 dopamine receptor binding / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / positive regulation of chemokine production / adenylate cyclase-activating adrenergic receptor signaling pathway / regulation of insulin secretion / cellular response to glucagon stimulus / presynaptic modulation of chemical synaptic transmission / adenylate cyclase activator activity / trans-Golgi network membrane / negative regulation of inflammatory response to antigenic stimulus / electron transport chain / bone development / G protein-coupled receptor activity / positive regulation of interleukin-6 production / platelet aggregation / vasodilation / cognition / Schaffer collateral - CA1 synapse / G-protein beta/gamma-subunit complex binding / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / sensory perception of smell / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / positive regulation of cold-induced thermogenesis / presynapse / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / G protein activity / GTPase binding / Ca2+ pathway / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / electron transfer activity / periplasmic space / Extra-nuclear estrogen signaling / cell population proliferation / iron ion binding / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / heme binding / synapse / GTP binding / protein-containing complex binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Oplophorus gracilirostris (crustacean) Oplophorus gracilirostris (crustacean) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.99 Å | |||||||||
 Authors Authors | Zhang JY / Chen Y / Hua T / Song GJ | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Cryo-EM structure of the human adenosine A receptor-G signaling complex. Authors: Ying Chen / Jinyi Zhang / Yuan Weng / Yueming Xu / Weiqiang Lu / Wei Liu / Mingyao Liu / Tian Hua / Gaojie Song /   Abstract: The human adenosine A receptor (AR) is a class A G protein-coupled receptor that is involved in several major physiological and pathological processes throughout the body. AR recognizes its ligands ...The human adenosine A receptor (AR) is a class A G protein-coupled receptor that is involved in several major physiological and pathological processes throughout the body. AR recognizes its ligands adenosine and NECA with relatively low affinity, but the detailed mechanism for its ligand recognition and signaling is still elusive. Here, we present two structures determined by cryo-electron microscopy of AR bound to its agonists NECA and BAY60-6583, each coupled to an engineered G protein. The structures reveal conserved orthosteric binding pockets with subtle differences, whereas the selectivity or specificity can mainly be attributed to regions extended from the orthosteric pocket. We also found that BAY60-6583 occupies a secondary pocket, where residues V250 and N273 were two key determinants for its selectivity against AR. This study offers a better understanding of ligand selectivity for the adenosine receptor family and provides a structural template for further development of AR ligands for related diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33512.map.gz emd_33512.map.gz | 57 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33512-v30.xml emd-33512-v30.xml emd-33512.xml emd-33512.xml | 24.8 KB 24.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33512.png emd_33512.png | 78.9 KB | ||
| Filedesc metadata |  emd-33512.cif.gz emd-33512.cif.gz | 7.5 KB | ||
| Others |  emd_33512_half_map_1.map.gz emd_33512_half_map_1.map.gz emd_33512_half_map_2.map.gz emd_33512_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33512 http://ftp.pdbj.org/pub/emdb/structures/EMD-33512 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33512 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33512 | HTTPS FTP |
-Related structure data
| Related structure data |  7xy6MC  7xy7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33512.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33512.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.96 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_33512_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
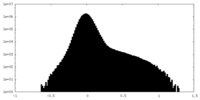
| Projections & Slices |
| ||||||||||||
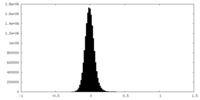
| Density Histograms |
-Half map: #2
| File | emd_33512_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
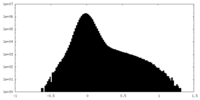
| Projections & Slices |
| ||||||||||||
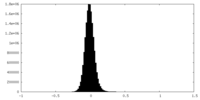
| Density Histograms |
- Sample components
Sample components
-Entire : Adenosine receptor bound to an agonist in complex with G protein ...
| Entire | Name: Adenosine receptor bound to an agonist in complex with G protein obtained by cryo-EM |
|---|---|
| Components |
|
-Supramolecule #1: Adenosine receptor bound to an agonist in complex with G protein ...
| Supramolecule | Name: Adenosine receptor bound to an agonist in complex with G protein obtained by cryo-EM type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: G protein
| Supramolecule | Name: G protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Oplophorus gracilirostris (crustacean) Oplophorus gracilirostris (crustacean) |
-Supramolecule #3: Soluble cytochrome b562,Adenosine receptor A2b,LgBiT
| Supramolecule | Name: Soluble cytochrome b562,Adenosine receptor A2b,LgBiT / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #4 |
|---|
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.146844 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TVSAEDKAAA ERSKMIEKQL QKDKQVYRAT HRLLLLGADN SGKSTIVKQM RIYHGGSGGS GGTSGIFETK FQVDKVNFHM FDVGGQRDE RRKWIQCFND VTAIIFVVDS SDYNRLQEAL NDFKSIWNNR WLRTISVILF LNKQDLLAEK VLAGKSKIED Y FPEFARYT ...String: TVSAEDKAAA ERSKMIEKQL QKDKQVYRAT HRLLLLGADN SGKSTIVKQM RIYHGGSGGS GGTSGIFETK FQVDKVNFHM FDVGGQRDE RRKWIQCFND VTAIIFVVDS SDYNRLQEAL NDFKSIWNNR WLRTISVILF LNKQDLLAEK VLAGKSKIED Y FPEFARYT TPEDATPEPG EDPRVTRAKY FIRDEFLRIS TASGDGRHYC YPHFTCAVDT ENARRIFNDC RDIIQRMHLR QY ELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.728426 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWNGSSGGG GSGGGGSSGV SGWRLFKKIS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.891022 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFGSA GSA |
-Macromolecule #4: Soluble cytochrome b562,Adenosine receptor A2b,LgBiT
| Macromolecule | Name: Soluble cytochrome b562,Adenosine receptor A2b,LgBiT / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Oplophorus gracilirostris (crustacean) Oplophorus gracilirostris (crustacean) |
| Molecular weight | Theoretical: 69.594055 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDAGR AHHHHHHHHH HADLEDNWET LNDNLKVIEK ADNAAQVKDA LTKMRAAALD AQKATPPKLE DKSPDSPEMK DFRHGFDIL VGQIDDALKL ANEGKVKEAQ AAAEQLKTTR NAYIQKYLEN LYFQSLLETQ DALYVALELV IAALSVAGNV L VCAAVGTA ...String: DYKDDDDAGR AHHHHHHHHH HADLEDNWET LNDNLKVIEK ADNAAQVKDA LTKMRAAALD AQKATPPKLE DKSPDSPEMK DFRHGFDIL VGQIDDALKL ANEGKVKEAQ AAAEQLKTTR NAYIQKYLEN LYFQSLLETQ DALYVALELV IAALSVAGNV L VCAAVGTA NTLQTPTNYF LVSLAAADVA VGLFAIPFAI TISLGFCTDF YGCLFLACFV LVLTQSSIFS LLAVAVDRYL AI CVPLRYK SLVTGTRARG VIAVLWVLAF GIGLTPFLGW NSKDSATNNC TEPWDGTTNE SCCLVKCLFE NVVPMSYMVY FNF FGCVLP PLLIMLVIYI KIFLVACRQL QRTELMDHSR TTLQREIHAA KSLAMIVGIF ALCWLPVHAV NCVTLFQPAQ GKNK PKWAM NMAILLSHAN SVVNPIVYAY RNRDFRYTFH KIISRYLLCQ ADVKSGNGQA GVQPALGVGL NSMGTLVFTL EDFVG DWEQ TAAYNLDQVL EQGGVSSLLQ NLAVSVTPIQ RIVRSGENAL KIDIHVIIPY EGLSADQMAQ IEEVFKVVYP VDDHHF KVI LPYGTLVIDG VTPNMLNYFG RPYEGIAVFD GKKITVTGTL WNGNKIIDER LITPDGSMLF RVTIN UniProtKB: Soluble cytochrome b562, Adenosine receptor A2b |
-Macromolecule #5: 2-[6-azanyl-3,5-dicyano-4-[4-(cyclopropylmethoxy)phenyl]pyridin-2...
| Macromolecule | Name: 2-[6-azanyl-3,5-dicyano-4-[4-(cyclopropylmethoxy)phenyl]pyridin-2-yl]sulfanylethanamide type: ligand / ID: 5 / Number of copies: 1 / Formula: I5D |
|---|---|
| Molecular weight | Theoretical: 379.436 Da |
| Chemical component information | 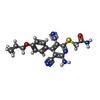 ChemComp-I5D: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)