+Search query
-Structure paper
| Title | Structural basis for broad coronavirus neutralization. |
|---|---|
| Journal, issue, pages | Nat Struct Mol Biol, Vol. 28, Issue 6, Page 478-486, Year 2021 |
| Publish date | May 12, 2021 |
 Authors Authors | Maximilian M Sauer / M Alejandra Tortorici / Young-Jun Park / Alexandra C Walls / Leah Homad / Oliver J Acton / John E Bowen / Chunyan Wang / Xiaoli Xiong / Willem de van der Schueren / Joel Quispe / Benjamin G Hoffstrom / Berend-Jan Bosch / Andrew T McGuire / David Veesler /     |
| PubMed Abstract | Three highly pathogenic β-coronaviruses have crossed the animal-to-human species barrier in the past two decades: SARS-CoV, MERS-CoV and SARS-CoV-2. To evaluate the possibility of identifying ...Three highly pathogenic β-coronaviruses have crossed the animal-to-human species barrier in the past two decades: SARS-CoV, MERS-CoV and SARS-CoV-2. To evaluate the possibility of identifying antibodies with broad neutralizing activity, we isolated a monoclonal antibody, termed B6, that cross-reacts with eight β-coronavirus spike glycoproteins, including all five human-infecting β-coronaviruses. B6 broadly neutralizes entry of pseudotyped viruses from lineages A and C, but not from lineage B, and the latter includes SARS-CoV and SARS-CoV-2. Cryo-EM, X-ray crystallography and membrane fusion assays reveal that B6 binds to a conserved cryptic epitope located in the fusion machinery. The data indicate that antibody binding sterically interferes with the spike conformational changes leading to membrane fusion. Our data provide a structural framework explaining B6 cross-reactivity with β-coronaviruses from three lineages, along with a proof of concept for antibody-mediated broad coronavirus neutralization elicited through vaccination. This study unveils an unexpected target for next-generation structure-guided design of a pan-β-coronavirus vaccine. |
 External links External links |  Nat Struct Mol Biol / Nat Struct Mol Biol /  PubMed:33981021 PubMed:33981021 |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 1.4 - 4.7 Å |
| Structure data |  EMDB-23672: EMDB-23674, PDB-7m5e:  PDB-7m51:  PDB-7m52:  PDB-7m53:  PDB-7m55: |
| Chemicals |  ChemComp-GOL:  ChemComp-HOH: 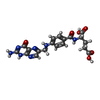 ChemComp-FOL:  ChemComp-NAG:  ChemComp-SIA: |
| Source |
|
 Keywords Keywords | ANTIVIRAL PROTEIN / IMMUNE SYSTEM / Broadly neutralizing antibody / Structural genomics / SSGCID / Seattle Structural Genomics Center for Infectious Disease / Center for Structural Genomics of Infectious Diseases / CSGID / VIRAL PROTEIN / MERS-CoV / coronaviruses / antibody |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers







