+Search query
-Structure paper
| Title | Discovery and Characterization of Clinical Candidate LXE408 as a Kinetoplastid-Selective Proteasome Inhibitor for the Treatment of Leishmaniases. |
|---|---|
| Journal, issue, pages | J Med Chem, Vol. 63, Issue 19, Page 10773-10781, Year 2020 |
| Publish date | Oct 8, 2020 |
 Authors Authors | Advait Nagle / Agnes Biggart / Celine Be / Honnappa Srinivas / Andreas Hein / Diana Caridha / Richard J Sciotti / Brandon Pybus / Mara Kreishman-Deitrick / Badry Bursulaya / Yin H Lai / Mu-Yun Gao / Fang Liang / Casey J N Mathison / Xiaodong Liu / Vince Yeh / Jeffrey Smith / Isabelle Lerario / Yongping Xie / Donatella Chianelli / Michael Gibney / Ashley Berman / Yen-Liang Chen / Jan Jiricek / Lauren C Davis / Xianzhong Liu / Jaime Ballard / Shilpi Khare / Fabian Kurt Eggimann / Alexandre Luneau / Todd Groessl / Michael Shapiro / Wendy Richmond / Kevin Johnson / Patrick J Rudewicz / Srinivasa P S Rao / Christopher Thompson / Tove Tuntland / Glen Spraggon / Richard J Glynne / Frantisek Supek / Christian Wiesmann / Valentina Molteni /   |
| PubMed Abstract | Visceral leishmaniasis is responsible for up to 30,000 deaths every year. Current treatments have shortcomings that include toxicity and variable efficacy across endemic regions. Previously, we ...Visceral leishmaniasis is responsible for up to 30,000 deaths every year. Current treatments have shortcomings that include toxicity and variable efficacy across endemic regions. Previously, we reported the discovery of GNF6702, a selective inhibitor of the kinetoplastid proteasome, which cleared parasites in murine models of leishmaniasis, Chagas disease, and human African trypanosomiasis. Here, we describe the discovery and characterization of LXE408, a structurally related kinetoplastid-selective proteasome inhibitor currently in Phase 1 human clinical trials. Furthermore, we present high-resolution cryo-EM structures of the proteasome in complex with LXE408, which provides a compelling explanation for the noncompetitive mode of binding of this novel class of inhibitors of the kinetoplastid proteasome. |
 External links External links |  J Med Chem / J Med Chem /  PubMed:32667203 / PubMed:32667203 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.2 - 3.4 Å |
| Structure data | EMDB-10462, PDB-6tcz: EMDB-10463, PDB-6td5: |
| Chemicals | 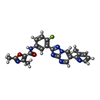 ChemComp-N2E: 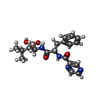 ChemComp-BO2: |
| Source |
|
 Keywords Keywords | HYDROLASE / Proteasome complex / inhibitor / peptidase |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers







 leishmania donovani (eukaryote)
leishmania donovani (eukaryote)