+Search query
-Structure paper
| Title | Conformational States of ABC Transporter MsbA in a Lipid Environment Investigated by Small-Angle Scattering Using Stealth Carrier Nanodiscs. |
|---|---|
| Journal, issue, pages | Structure, Vol. 26, Issue 8, Page 1072-11079.e4, Year 2018 |
| Publish date | Aug 7, 2018 |
 Authors Authors | Inokentijs Josts / Julius Nitsche / Selma Maric / Haydyn D Mertens / Martine Moulin / Michael Haertlein / Sylvain Prevost / Dmitri I Svergun / Sebastian Busch / V Trevor Forsyth / Henning Tidow /     |
| PubMed Abstract | Structural studies of integral membrane proteins (IMPs) are challenging, as many of them are inactive or insoluble in the absence of a lipid environment. Here, we describe an approach making use of ...Structural studies of integral membrane proteins (IMPs) are challenging, as many of them are inactive or insoluble in the absence of a lipid environment. Here, we describe an approach making use of fractionally deuterium labeled "stealth carrier" nanodiscs that are effectively invisible to low-resolution neutron diffraction and enable structural studies of IMPs in a lipidic native-like solution environment. We illustrate the potential of the method in a joint small-angle neutron scattering (SANS) and X-ray scattering (SAXS) study of the ATP-binding cassette (ABC) transporter protein MsbA solubilized in the stealth nanodiscs. The data allow for a direct observation of the signal from the solubilized protein without contribution from the surrounding lipid nanodisc. Not only the overall shape but also differences between conformational states of MsbA can be reliably detected from the scattering data, demonstrating the sensitivity of the approach and its general applicability to structural studies of IMPs. |
 External links External links |  Structure / Structure /  PubMed:29937358 PubMed:29937358 |
| Methods | SAS (neutron source) / SAS (X-ray synchrotron) |
| Structure data |  SASDDB5:  SASDDC5: MsbA in stealth nanodisc (SANS, 100% D2O) + 1 mM ADP  SASDDD5: MsbA in stealth nanodisc (SAXS) (Lipid A export ATP-binding/permease protein MsbA, MsbA + Membrane scaffold protein 1D1 (deuterated, 75%), d75-msp1d1 + 1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl), d(78,92)-POPC) 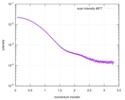 SASDDE5: NBD-MsbA (apo) (Nucleotide Binding Domain of Lipid A export ATP-binding/permease protein MsbA, NBD-MsbA)  SASDDF5: NBD-MsbA (+1mM ADP) (Nucleotide Binding Domain of Lipid A export ATP-binding/permease protein MsbA, NBD-MsbA) |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




