+Search query
-Structure paper
| Title | Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis. |
|---|---|
| Journal, issue, pages | Nucleic Acids Res, Vol. 45, Issue 18, Page 10884-10894, Year 2017 |
| Publish date | Oct 13, 2017 |
 Authors Authors | Kailu Yang / Jeng-Yih Chang / Zhicheng Cui / Xiaojun Li / Ran Meng / Lijun Duan / Jirapat Thongchol / Joanita Jakana / Christoph M Huwe / James C Sacchettini / Junjie Zhang /   |
| PubMed Abstract | Ribosomes from Mycobacterium tuberculosis (Mtb) possess species-specific ribosomal RNA (rRNA) expansion segments and ribosomal proteins (rProtein). Here, we present the near-atomic structures of the ...Ribosomes from Mycobacterium tuberculosis (Mtb) possess species-specific ribosomal RNA (rRNA) expansion segments and ribosomal proteins (rProtein). Here, we present the near-atomic structures of the Mtb 50S ribosomal subunit and the complete Mtb 70S ribosome, solved by cryo-electron microscopy. Upon joining of the large and small ribosomal subunits, a 100-nt long expansion segment of the Mtb 23S rRNA, named H54a or the 'handle', switches interactions from with rRNA helix H68 and rProtein uL2 to with rProtein bS6, forming a new intersubunit bridge 'B9'. In Mtb 70S, bridge B9 is mostly maintained, leading to correlated motions among the handle, the L1 stalk and the small subunit in the rotated and non-rotated states. Two new protein densities were discovered near the decoding center and the peptidyl transferase center, respectively. These results provide a structural basis for studying translation in Mtb as well as developing new tuberculosis drugs. |
 External links External links |  Nucleic Acids Res / Nucleic Acids Res /  PubMed:28977617 / PubMed:28977617 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.7 - 7.1 Å |
| Structure data | EMDB-8641: Cryo-EM structure of the large ribosomal subunit from Mycobacterium tuberculosis EMDB-8645, PDB-5v93:  EMDB-8646:  EMDB-8647:  EMDB-8648:  EMDB-8649: |
| Chemicals | 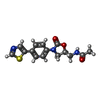 ChemComp-917: |
| Source |
|
 Keywords Keywords | RIBOSOME / RNA dynamics / RNA complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers








