+Search query
-Structure paper
| Title | Negative Stain Single-particle EM of the Maltose Transporter in Nanodiscs Reveals Asymmetric Closure of MalK and Catalytic Roles of ATP, MalE, and Maltose. |
|---|---|
| Journal, issue, pages | J Biol Chem, Vol. 292, Issue 13, Page 5457-5464, Year 2017 |
| Publish date | Mar 31, 2017 |
 Authors Authors | Lucien Fabre / Huan Bao / James Innes / Franck Duong / Isabelle Rouiller /  |
| PubMed Abstract | The MalE-MalFGK complex is one of the best characterized members of the large and ubiquitous family of ATP-binding cassette (ABC) transporters. It is composed of a membrane-spanning heterodimer, ...The MalE-MalFGK complex is one of the best characterized members of the large and ubiquitous family of ATP-binding cassette (ABC) transporters. It is composed of a membrane-spanning heterodimer, MalF-MalG; a homodimeric ATPase, MalK; and a periplasmic maltose receptor, MalE. Opening and closure of MalK is coupled to conformational changes in MalF-MalG and the alternate exposition of the substrate-binding site to either side of the membrane. To further define this alternate access mechanism and the impact of ATP, MalE, and maltose on the conformation of the transporter during the transport cycle, we have reconstituted MalFGK in nanodiscs and analyzed its conformations under 10 different biochemical conditions using negative stain single-particle EM. EM map results (at 15-25 Å resolution) indicate that binding of ATP to MalK promotes an asymmetric, semi-closed conformation in accordance with the low ATPase activity of MalFGK In the presence of MalE, the MalK dimer becomes fully closed, gaining the ability to hydrolyze ATP. In the presence of ADP or maltose, MalE·MalFGK remains essentially in a semi-closed symmetric conformation, indicating that release of these ligands is required for the return to the initial state. Taken together, this structural information provides a rationale for the stimulation of MalK ATPase activity by MalE as well as by maltose. |
 External links External links |  J Biol Chem / J Biol Chem /  PubMed:28188291 / PubMed:28188291 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 18.8 - 25.2 Å |
| Structure data |  EMDB-8524:  EMDB-8525:  EMDB-8526:  EMDB-8527: 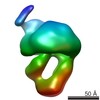 EMDB-8529:  EMDB-8530: 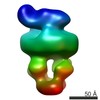 EMDB-8531: 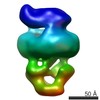 EMDB-8533:  EMDB-8534:  EMDB-8535: 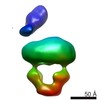 EMDB-8536: |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




