+Search query
-Structure paper
| Title | Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis. |
|---|---|
| Journal, issue, pages | Mol. Microbiol., Vol. 103, Page 1034-1045, Year 2017 |
| Publish date | Dec 19, 2016 (structure data deposition date) |
 Authors Authors | Molodtsov, V. / Scharf, N.T. / Stefan, M.A. / Garcia, G.A. / Murakami, K.S. |
 External links External links |  Mol. Microbiol. / Mol. Microbiol. /  PubMed:28009073 PubMed:28009073 |
| Methods | X-ray diffraction |
| Resolution | 3.399 - 4.1 Å |
| Structure data |  PDB-5uac:  PDB-5uag:  PDB-5uah:  PDB-5uaj:  PDB-5ual:  PDB-5uaq: |
| Chemicals | 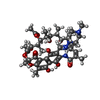 ChemComp-RFP:  ChemComp-MG:  ChemComp-ZN: |
| Source |
|
 Keywords Keywords | transferase/transferase inhibitor / inhibitor / antibiotic / tuberculosis / rifampin / rifamycin / RNA polymerase / transferase-transferase inhibitor complex / rifamycin resistant mutation / rpoB-D516V / RpoB D516V mutant / RpoB S531L / TRANSFERASE / RpoB H526Y |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




