+Search query
-Structure paper
| Title | MuB is an AAA+ ATPase that forms helical filaments to control target selection for DNA transposition. |
|---|---|
| Journal, issue, pages | Proc Natl Acad Sci U S A, Vol. 110, Issue 27, Page E2441-E2450, Year 2013 |
| Publish date | Jul 2, 2013 |
 Authors Authors | Naoko Mizuno / Marija Dramićanin / Michiyo Mizuuchi / Julia Adam / Yi Wang / Yong-Woon Han / Wei Yang / Alasdair C Steven / Kiyoshi Mizuuchi / Santiago Ramón-Maiques /  |
| PubMed Abstract | MuB is an ATP-dependent nonspecific DNA-binding protein that regulates the activity of the MuA transposase and captures target DNA for transposition. Mechanistic understanding of MuB function has ...MuB is an ATP-dependent nonspecific DNA-binding protein that regulates the activity of the MuA transposase and captures target DNA for transposition. Mechanistic understanding of MuB function has previously been hindered by MuB's poor solubility. Here we combine bioinformatic, mutagenic, biochemical, and electron microscopic analyses to unmask the structure and function of MuB. We demonstrate that MuB is an ATPase associated with diverse cellular activities (AAA+ ATPase) and forms ATP-dependent filaments with or without DNA. We also identify critical residues for MuB's ATPase, DNA binding, protein polymerization, and MuA interaction activities. Using single-particle electron microscopy, we show that MuB assembles into a helical filament, which binds the DNA in the axial channel. The helical parameters of the MuB filament do not match those of the coated DNA. Despite this protein-DNA symmetry mismatch, MuB does not deform the DNA duplex. These findings, together with the influence of MuB filament size on strand-transfer efficiency, lead to a model in which MuB-imposed symmetry transiently deforms the DNA at the boundary of the MuB filament and results in a bent DNA favored by MuA for transposition. |
 External links External links |  Proc Natl Acad Sci U S A / Proc Natl Acad Sci U S A /  PubMed:23776210 / PubMed:23776210 /  PubMed Central PubMed Central |
| Methods | EM (helical sym.) / EM (single particle) |
| Resolution | 16.0 - 18.0 Å |
| Structure data | EMDB-2395: MuB is an AAA+ ATPase that forms helical filaments to control target selection for DNA transposition |
| Chemicals |  ChemComp-ADP: |
| Source |
|
 Keywords Keywords | HYDROLASE / AAA+ DNA TRANSPOSITION / NUCLEOPROTEIN FILAMENT / SYMMETRY MISMATCH / TRANSCRIPTION / AAA+ ATPASE / DNA TRANSPOSITION / AAAPLUS ATPASE |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



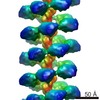



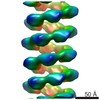

 enterobacteria phage mu (virus)
enterobacteria phage mu (virus)