-Search query
-Search result
Showing 1 - 50 of 343 items for (author: chou & t)

EMDB-38216: 
Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with antibody O5C2

PDB-8xbf: 
Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with antibody O5C2
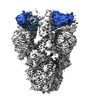
EMDB-39546: 
SARS-CoV-2 Delta Spike in complex with JL-8C

EMDB-39547: 
SARS-CoV-2 Delta Spike in complex with JM-1A

EMDB-39685: 
SARS-CoV-2 Delta Spike in complex with Fab of JE-5C

EMDB-39686: 
SARS-CoV-2 Spike (BA.1) in complex with Fab of JH-8B

PDB-8yro: 
SARS-CoV-2 Delta Spike in complex with JL-8C

PDB-8yrp: 
SARS-CoV-2 Delta Spike in complex with JM-1A

PDB-8yz5: 
SARS-CoV-2 Delta Spike in complex with Fab of JE-5C

PDB-8yz6: 
SARS-CoV-2 Spike (BA.1) in complex with Fab of JH-8B
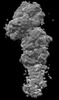
EMDB-44479: 
Cryo-EM structure of synthetic claudin-4 complex with Clostridium perfringens enterotoxin C-terminal domain, sFab COP-2, and Nanobody

PDB-9bei: 
Cryo-EM structure of synthetic claudin-4 complex with Clostridium perfringens enterotoxin C-terminal domain, sFab COP-2, and Nanobody

EMDB-36800: 
Potassium transporter KtrAB from Bacillus subtilis in ADP-bound state

EMDB-36801: 
Potassium transporter KtrAB from Bacillus subtilis in ADP-bound state, focused refined on KtrA octamer

EMDB-36802: 
Potassium transporter KtrAB from Bacillus subtilis in ADP-bound state, focused refined on KtrB dimer

EMDB-36803: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of MgCl2

EMDB-36804: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA

EMDB-38477: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, vertical C2 symmetry axis

EMDB-38478: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, C1 symmetry

PDB-8k1s: 
Potassium transporter KtrAB from Bacillus subtilis in ADP-bound state

PDB-8k1t: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of MgCl2

PDB-8k1u: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA

PDB-8xmh: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, vertical C2 symmetry axis

PDB-8xmi: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, C1 symmetry

EMDB-43658: 
SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs

EMDB-43659: 
SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement

EMDB-43660: 
SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement

PDB-8vye: 
SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs

PDB-8vyf: 
SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement

PDB-8vyg: 
SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement

EMDB-41423: 
Cryo-EM structure of DDB1dB:CRBN:Pomalidomide:SD40

EMDB-41424: 
Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 1

EMDB-41425: 
Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 2

EMDB-41777: 
Map from local refinement (focused on CRBN) of DDB1dB:CRBN:Pomalidomide:SD40

EMDB-41778: 
Map from local refinement (focused on CRBN) of DDB1dB:CRBN:PT-179:SD40, conformation 1

EMDB-41779: 
Map from local refinement (focused on CRBN) of DDB1dB:CRBN:PT-179:SD40, conformation 2

PDB-8tnp: 
Cryo-EM structure of DDB1dB:CRBN:Pomalidomide:SD40

PDB-8tnq: 
Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 1

PDB-8tnr: 
Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 2

EMDB-19431: 
Cryo-em structure of the rat Multidrug resistance-associated protein 2 (rMrp2) in an autoinhibited state (nucleotide-free)

EMDB-19433: 
Cryo-em structure of the rat Multidrug resistance-associated protein 2 (rMrp2) in complex with probenecid

PDB-8rq3: 
Cryo-em structure of the rat Multidrug resistance-associated protein 2 (rMrp2) in an autoinhibited state (nucleotide-free)

PDB-8rq4: 
Cryo-em structure of the rat Multidrug resistance-associated protein 2 (rMrp2) in complex with probenecid

EMDB-40811: 
Cryo-EM structure of NLRP3 open octamer
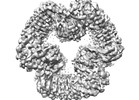
EMDB-40820: 
Cryo-EM structure of NLRP3 closed hexamer
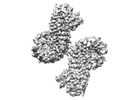
EMDB-40855: 
Structure of NLRP3 and NEK7 complex

PDB-8swf: 
Cryo-EM structure of NLRP3 open octamer

PDB-8swk: 
Cryo-EM structure of NLRP3 closed hexamer

PDB-8sxn: 
Structure of NLRP3 and NEK7 complex

EMDB-42787: 
Arp2/3 branch junction complex, ADP state
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
