[English] 日本語
 Yorodumi
Yorodumi- EMDB-8695: BG505 SOSIP.664 in complex with broadly neutralizing antibodies P... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8695 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
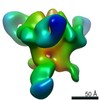
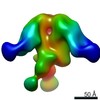
| Title | BG505 SOSIP.664 in complex with broadly neutralizing antibodies PG9 and 8ANC195 | |||||||||
 Map data Map data | BG505 SOSIP-PG9-8ANC195 | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information immunoglobulin complex / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / immunoglobulin complex / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell /  adaptive immune response / adaptive immune response /  viral protein processing / fusion of virus membrane with host plasma membrane ... viral protein processing / fusion of virus membrane with host plasma membrane ... immunoglobulin complex / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / immunoglobulin complex / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell /  adaptive immune response / adaptive immune response /  viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / extracellular region / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / extracellular region /  membrane / identical protein binding / membrane / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /    Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
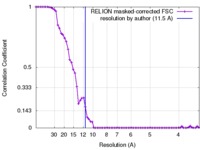
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 11.5 Å cryo EM / Resolution: 11.5 Å | |||||||||
 Authors Authors | Wang H / Bjorkman PJ | |||||||||
 Citation Citation |  Journal: Elife / Year: 2017 Journal: Elife / Year: 2017Title: Asymmetric recognition of HIV-1 Envelope trimer by V1V2 loop-targeting antibodies. Authors: Haoqing Wang / Harry B Gristick / Louise Scharf / Anthony P West / Rachel P Galimidi / Michael S Seaman / Natalia T Freund / Michel C Nussenzweig / Pamela J Bjorkman /  Abstract: The HIV-1 envelope (Env) glycoprotein binds to host cell receptors to mediate membrane fusion. The prefusion Env trimer is stabilized by V1V2 loops that interact at the trimer apex. Broadly ...The HIV-1 envelope (Env) glycoprotein binds to host cell receptors to mediate membrane fusion. The prefusion Env trimer is stabilized by V1V2 loops that interact at the trimer apex. Broadly neutralizing antibodies (bNAbs) against V1V2 loops, exemplified by PG9, bind asymmetrically as a single Fab to the apex of the symmetric Env trimer using a protruding CDRH3 to penetrate the Env glycan shield. Here we characterized a distinct mode of V1V2 epitope recognition by the new bNAb BG1 in which two Fabs bind asymmetrically per Env trimer using a compact CDRH3. Comparisons between cryo-EM structures of Env trimer complexed with BG1 (6.2 Å resolution) and PG9 (11.5 Å resolution) revealed a new V1V2-targeting strategy by BG1. Analyses of the EM structures provided information relevant to vaccine design including molecular details for different modes of asymmetric recognition of Env trimer and a binding model for BG1 recognition of V1V2 involving glycan flexibility. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8695.map.gz emd_8695.map.gz | 24.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8695-v30.xml emd-8695-v30.xml emd-8695.xml emd-8695.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8695_fsc.xml emd_8695_fsc.xml | 6.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_8695.png emd_8695.png | 82 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8695 http://ftp.pdbj.org/pub/emdb/structures/EMD-8695 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8695 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8695 | HTTPS FTP |
-Related structure data
| Related structure data |  5vj6MC  8693C  5viyC  5vvfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8695.map.gz / Format: CCP4 / Size: 26.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8695.map.gz / Format: CCP4 / Size: 26.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BG505 SOSIP-PG9-8ANC195 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.75 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : BG505 SOSIP-PG9-8ANC195 complex
| Entire | Name: BG505 SOSIP-PG9-8ANC195 complex |
|---|---|
| Components |
|
-Supramolecule #1: BG505 SOSIP-PG9-8ANC195 complex
| Supramolecule | Name: BG505 SOSIP-PG9-8ANC195 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.146482 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEA QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD |
-Macromolecule #2: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 54.064277 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT ...String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT SAITQACPKV SFEPIPIHYC APAGFAILKC KDKKFNGTGP CPSVSTVQCT HGIKPVVSTQ LLLNGSLAEE EV MIRSENI TNNAKNILVQ FNTPVQINCT RPNNNTRKSI RIGPGQAFYA TGDIIGDIRQ AHCNVSKATW NETLGKVVKQ LRK HFGNNT IIRFANSSGG DLEVTTHSFN CGGEFFYCNT SGLFNSTWIS NTSVQGSNST GSNDSITLPC RIKQIINMWQ RIGQ AMYAP PIQGVIRCVS NITGLILTRD GGSTNSTTET FRPGGGDMRD NWRSELYKYK VVKIEPLGVA PTRCKRRVVG RRRRR R |
-Macromolecule #3: PG9 Fab heavy chain
| Macromolecule | Name: PG9 Fab heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 27.211188 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ERLVESGGGV VQPGSSLRLS CAASGFDFSR QGMHWVRQAP GQGLEWVAFI KYDGSEKYHA DSVWGRLSIS RDNSKDTLYL QMNSLRVED TATYFCVREA GGPDYRNGYN (TYS)(TYS)DFYDGYYN YHYMDVWGKG TTVTVSSAST KGPSVFPLAP SSK STSGGT ...String: ERLVESGGGV VQPGSSLRLS CAASGFDFSR QGMHWVRQAP GQGLEWVAFI KYDGSEKYHA DSVWGRLSIS RDNSKDTLYL QMNSLRVED TATYFCVREA GGPDYRNGYN (TYS)(TYS)DFYDGYYN YHYMDVWGKG TTVTVSSAST KGPSVFPLAP SSK STSGGT AALGCLVKDY FPEPVTVSWN SGALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KVDK KVEPK SCDKGLEVLF Q |
-Macromolecule #4: 8ANC195 Fab heavy chain
| Macromolecule | Name: 8ANC195 Fab heavy chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.788809 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QIHLVQSGTE VKKPGSSVTV SCKAYGVNTF GLYAVNWVRQ APGQSLEYIG QIWRWKSSAS HHFRGRVLIS AVDLTGSSPP ISSLEIKNL TSDDTAVYFC TTTSTYDRWS GLHHDGVMAF SSWGQGTLIS VSAASTKGPS VFPLAPSSKS TSGGTAALGC L VKDYFPEP ...String: QIHLVQSGTE VKKPGSSVTV SCKAYGVNTF GLYAVNWVRQ APGQSLEYIG QIWRWKSSAS HHFRGRVLIS AVDLTGSSPP ISSLEIKNL TSDDTAVYFC TTTSTYDRWS GLHHDGVMAF SSWGQGTLIS VSAASTKGPS VFPLAPSSKS TSGGTAALGC L VKDYFPEP VTVSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK RVEPK |
-Macromolecule #5: 8ANC195 Fab light chain
| Macromolecule | Name: 8ANC195 Fab light chain / type: protein_or_peptide / ID: 5 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.460047 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPST LSASTGDTVR ISCRASQSIT GNWVAWYQQR PGKAPRLLIY RGAALLGGVP SRFRGSAAGT DFTLTIGNLQ AEDFGTFYC QQYDTYPGTF GQGTKVEVKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: DIQMTQSPST LSASTGDTVR ISCRASQSIT GNWVAWYQQR PGKAPRLLIY RGAALLGGVP SRFRGSAAGT DFTLTIGNLQ AEDFGTFYC QQYDTYPGTF GQGTKVEVKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #6: PG9 Fab light chain
| Macromolecule | Name: PG9 Fab light chain / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.837256 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSALTQPASV SGSPGQSITI SCQGTSNDVG GYESVSWYQQ HPGKAPKVVI YDVSKRPSGV SNRFSGSKSG NTASLTISGL QAEDEGDYY CKSLTSTRRR VFGTGTKLTV LGQPKAAPSV TLFPPSSEEL QANKATLVCL ISDFYPGAVT VAWKADSSPV K AGVETTTP ...String: QSALTQPASV SGSPGQSITI SCQGTSNDVG GYESVSWYQQ HPGKAPKVVI YDVSKRPSGV SNRFSGSKSG NTASLTISGL QAEDEGDYY CKSLTSTRRR VFGTGTKLTV LGQPKAAPSV TLFPPSSEEL QANKATLVCL ISDFYPGAVT VAWKADSSPV K AGVETTTP SKQSNNKYAA SSYLSLTPEQ WKSHKSYSCQ VTHEGSTVEK TVAPTECS |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 30.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-5vj6: |
 Movie
Movie Controller
Controller