+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3771 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Three-dimensional reconstruction of FLiP virion | |||||||||
 Map data Map data | Reconstruction of Flavobacterium infecting lipid-containing phage FLiP | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | Major capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |   unidentified phage (virus) unidentified phage (virus) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.0 Å cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | De Colibus L / Gillum A / Stuart DI / Huiskonen JT | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2017 Journal: Proc Natl Acad Sci U S A / Year: 2017Title: Virus found in a boreal lake links ssDNA and dsDNA viruses. Authors: Elina Laanto / Sari Mäntynen / Luigi De Colibus / Jenni Marjakangas / Ashley Gillum / David I Stuart / Janne J Ravantti / Juha T Huiskonen / Lotta-Riina Sundberg /   Abstract: Viruses have impacted the biosphere in numerous ways since the dawn of life. However, the evolution, genetic, structural, and taxonomic diversity of viruses remain poorly understood, in part because ...Viruses have impacted the biosphere in numerous ways since the dawn of life. However, the evolution, genetic, structural, and taxonomic diversity of viruses remain poorly understood, in part because sparse sampling of the virosphere has concentrated mostly on exploring the abundance and diversity of dsDNA viruses. Furthermore, viral genomes are highly diverse, and using only the current sequence-based methods for classifying viruses and studying their phylogeny is complicated. Here we describe a virus, FLiP (-infecting, lipid-containing phage), with a circular ssDNA genome and an internal lipid membrane enclosed in the icosahedral capsid. The 9,174-nt-long genome showed limited sequence similarity to other known viruses. The genetic data imply that this virus might use replication mechanisms similar to those found in other ssDNA replicons. However, the structure of the viral major capsid protein, elucidated at near-atomic resolution using cryo-electron microscopy, is strikingly similar to that observed in dsDNA viruses of the PRD1-adenovirus lineage, characterized by a major capsid protein bearing two β-barrels. The strong similarity between FLiP and another member of the structural lineage, bacteriophage PM2, extends to the capsid organization (pseudo = 21 ) despite the difference in the genetic material packaged and the lack of significant sequence similarity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3771.map.gz emd_3771.map.gz | 1.2 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3771-v30.xml emd-3771-v30.xml emd-3771.xml emd-3771.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3771_fsc.xml emd_3771_fsc.xml | 24.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_3771.png emd_3771.png | 199.4 KB | ||
| Masks |  emd_3771_msk_1.map emd_3771_msk_1.map | 1.3 GB |  Mask map Mask map | |
| Others |  emd_3771_half_map_1.map.gz emd_3771_half_map_1.map.gz emd_3771_half_map_2.map.gz emd_3771_half_map_2.map.gz | 1 GB 1 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3771 http://ftp.pdbj.org/pub/emdb/structures/EMD-3771 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3771 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3771 | HTTPS FTP |
-Related structure data
| Related structure data |  5oacMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3771.map.gz / Format: CCP4 / Size: 1.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3771.map.gz / Format: CCP4 / Size: 1.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of Flavobacterium infecting lipid-containing phage FLiP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
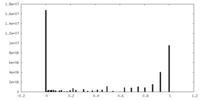
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_3771_msk_1.map emd_3771_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
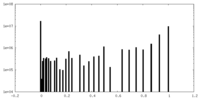
| Density Histograms |
-Half map: #1
| File | emd_3771_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
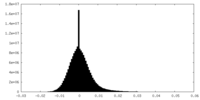
| Density Histograms |
-Half map: #2
| File | emd_3771_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
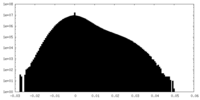
| Density Histograms |
- Sample components
Sample components
-Entire : unidentified phage
| Entire | Name:   unidentified phage (virus) unidentified phage (virus) |
|---|---|
| Components |
|
-Supramolecule #1: unidentified phage
| Supramolecule | Name: unidentified phage / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Flavobacterium infecting lipid-containing phage FLiP NCBI-ID: 38018 / Sci species name: unidentified phage / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:   Flavobacterium (bacteria) / Strain: sp B330 Flavobacterium (bacteria) / Strain: sp B330 |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 550.0 Å / T number (triangulation number): 25 |
-Macromolecule #1: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   unidentified phage (virus) unidentified phage (virus) |
| Molecular weight | Theoretical: 34.533934 KDa |
| Sequence | String: MTIKYLSSET EKLMNQTVSG IDVCFTLIGV DDDSFASGSK NDYISDTPKF LDPSNVHIKA TLKRGGKDYV LFSENLALLA KYSTITQGR DQWEEGVKLA AKEMVHLVYI PFSGNTNWPA HINLKDNDVL EVYVNVVRGA YGAELDANAC ICDVRTSPSI G VEKFIPFM ...String: MTIKYLSSET EKLMNQTVSG IDVCFTLIGV DDDSFASGSK NDYISDTPKF LDPSNVHIKA TLKRGGKDYV LFSENLALLA KYSTITQGR DQWEEGVKLA AKEMVHLVYI PFSGNTNWPA HINLKDNDVL EVYVNVVRGA YGAELDANAC ICDVRTSPSI G VEKFIPFM TSYSIRANQA TDLVNLGNDV TRIALLSMTN DVSNIPNAFT DVTLSSDRLD KNFNSNQLIL EHSKCIEDSV RS HANEVDS YLIHEDIEID SAKVHLKMNP AKIRENTIYL VRSHFQTSLE ILQKAVAMEE KHQSADIAKV PAT |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 / Details: 20 mM PBS |
|---|---|
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 0.7 µm / Calibrated magnification: 37037 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm / Nominal magnification: 160000 Bright-field microscopy / Cs: 2.0 mm / Nominal magnification: 160000 |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Sampling interval: 5.0 µm / Digitization - Frames/image: 1-22 / Average electron dose: 22.0 e/Å2 |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 84.31 / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model |  PDB-5oac: |
 Movie
Movie Controller
Controller












 Z
Z Y
Y X
X