[English] 日本語
 Yorodumi
Yorodumi- EMDB-3287: Evidence for a conformational switch in Influenzavirus M1 and its... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3287 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Evidence for a conformational switch in Influenzavirus M1 and its role in filamentous virion architecture | |||||||||
 Map data Map data | Reconstruction of trailing tips from influenza A virions. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Influenza A virus / FLUAV / Influenza A virus / FLUAV /  polymerase / M1 / crown / leading tip / trailing tip / polymerase / M1 / crown / leading tip / trailing tip /  filamentous filamentous | |||||||||
| Biological species |    Influenza A virus Influenza A virus | |||||||||
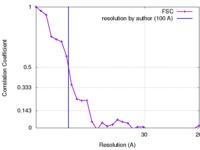
| Method | subtomogram averaging /  cryo EM / Resolution: 100.0 Å cryo EM / Resolution: 100.0 Å | |||||||||
 Authors Authors | Kiss G / Abdulsattar BO / Phapugrangkul P / Birch K / Jones IM / Neuman BW | |||||||||
 Citation Citation | Journal: Proc Natl Acad Sci U S A / Year: 2010 Title: Structural organization of a filamentous influenza A virus. Authors: Lesley J Calder / Sebastian Wasilewski / John A Berriman / Peter B Rosenthal /  Abstract: Influenza is a lipid-enveloped, pleomorphic virus. We combine electron cryotomography and analysis of images of frozen-hydrated virions to determine the structural organization of filamentous ...Influenza is a lipid-enveloped, pleomorphic virus. We combine electron cryotomography and analysis of images of frozen-hydrated virions to determine the structural organization of filamentous influenza A virus. Influenza A/Udorn/72 virions are capsule-shaped or filamentous particles of highly uniform diameter. We show that the matrix layer adjacent to the membrane is an ordered helix of the M1 protein and its close interaction with the surrounding envelope determines virion morphology. The ribonucleoprotein particles (RNPs) that package the genome segments form a tapered assembly at one end of the virus interior. The neuraminidase, which is present in smaller numbers than the hemagglutinin, clusters in patches and are typically present at the end of the virion opposite to RNP attachment. Incubation of virus at low pH causes a loss of filamentous morphology, during which we observe a structural transition of the matrix layer from its helical, membrane-associated form to a multilayered coil structure inside the virus particle. The polar organization of the virus provides a model for assembly of the virion during budding at the host membrane. Images and tomograms of A/Aichi/68 X-31 virions show the generality of these conclusions to non-filamentous virions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3287.map.gz emd_3287.map.gz | 854 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3287-v30.xml emd-3287-v30.xml emd-3287.xml emd-3287.xml | 9.4 KB 9.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3287_fsc.xml emd_3287_fsc.xml | 2.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_3287.png emd_3287.png | 74.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3287 http://ftp.pdbj.org/pub/emdb/structures/EMD-3287 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3287 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3287 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3287.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3287.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of trailing tips from influenza A virions. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 10 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Trailing tips of filamentous influenza A virions, showing M1 and ...
| Entire | Name: Trailing tips of filamentous influenza A virions, showing M1 and parts of the viral ribonucleoprotein |
|---|---|
| Components |
|
-Supramolecule #1000: Trailing tips of filamentous influenza A virions, showing M1 and ...
| Supramolecule | Name: Trailing tips of filamentous influenza A virions, showing M1 and parts of the viral ribonucleoprotein type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Influenza A virus
| Supramolecule | Name: Influenza A virus / type: virus / ID: 1 / Name.synonym: Influenza virus Details: Reconstruction of the inner layer of the viral envelope from trailing tips of native virions of mixed A/Aichi/X31 and A/Udorn strains. Map is Gaussian filtered to 7.6 nm the 0.5 FSC. Reconstructed in EMAN2. NCBI-ID: 11320 / Sci species name: Influenza A virus / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: No / Syn species name: Influenza virus |
|---|---|
| Host (natural) | Organism:   Homo sapiens (human) / Strain: Combined A/Aichi/2/68 and A/Udorn/307/72 / synonym: VERTEBRATES Homo sapiens (human) / Strain: Combined A/Aichi/2/68 and A/Udorn/307/72 / synonym: VERTEBRATES |
| Host system | Organism:   Homo sapiens (human) / Recombinant cell: MDCK Homo sapiens (human) / Recombinant cell: MDCK |
| Virus shell | Shell ID: 1 / Name: M1 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: FEI VITROBOT MARK III Details: Details for EM and reconstruction published in Calder et al., Proc Natl Acad Sci U S A. 2010 Jun 8; 107(23): 10685 to 10690. Method: Blotted 4s before plunging |
|---|
- Electron microscopy
Electron microscopy
| Microscope | OTHER |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Date | Jan 1, 2010 |
| Image recording | Category: CCD / Film or detector model: FEI EAGLE (2k x 2k) / Average electron dose: 50 e/Å2 |
 Movie
Movie Controller
Controller