+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1254 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
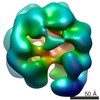
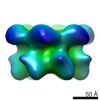
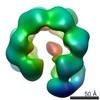
| Title | Hexameric ring structure of human MCM10 DNA replication factor. | |||||||||
 Map data Map data | Surface views of human Mcm10 top side bottom stereo | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 16.0 Å negative staining / Resolution: 16.0 Å | |||||||||
 Authors Authors | Okorokov AL / Orlova EV | |||||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2007 Journal: EMBO Rep / Year: 2007Title: Hexameric ring structure of human MCM10 DNA replication factor. Authors: Andrei L Okorokov / Alastair Waugh / Julie Hodgkinson / Andal Murthy / Hye Kyung Hong / Elisabetta Leo / Michael B Sherman / Kai Stoeber / Elena V Orlova / Gareth H Williams /  Abstract: The DNA replication factor minichromosome maintenance 10 (MCM10) is a conserved, abundant nuclear protein crucial for origin firing. During the transition from pre-replicative complexes to pre- ...The DNA replication factor minichromosome maintenance 10 (MCM10) is a conserved, abundant nuclear protein crucial for origin firing. During the transition from pre-replicative complexes to pre-initiation complexes, MCM10 recruitment to replication origins is required to provide a physical link between the MCM2-7 complex DNA helicase and DNA polymerases. Here, we report the molecular structure of human MCM10 as determined by electron microscopy and single-particle analysis. The MCM10 molecule is a ring-shaped hexamer with large central and smaller lateral channels and a system of inner chambers. This structure, together with biochemical data, suggests that this important protein uses its architecture to provide a docking module for assembly of the molecular machinery required for eukaryotic DNA replication. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1254.map.gz emd_1254.map.gz | 1.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1254-v30.xml emd-1254-v30.xml emd-1254.xml emd-1254.xml | 10.4 KB 10.4 KB | Display Display |  EMDB header EMDB header |
| Images |  1254.gif 1254.gif | 64.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1254 http://ftp.pdbj.org/pub/emdb/structures/EMD-1254 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1254 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1254 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1254.map.gz / Format: CCP4 / Size: 6.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1254.map.gz / Format: CCP4 / Size: 6.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Surface views of human Mcm10 top side bottom stereo | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.59 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : recombinant human Mcm10
| Entire | Name: recombinant human Mcm10 |
|---|---|
| Components |
|
-Supramolecule #1000: recombinant human Mcm10
| Supramolecule | Name: recombinant human Mcm10 / type: sample / ID: 1000 / Details: monodisperse / Oligomeric state: homohexamer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 650 KDa / Theoretical: 590 KDa / Method: size-exclusion FPLC |
-Macromolecule #1: Mcm10
| Macromolecule | Name: Mcm10 / type: protein_or_peptide / ID: 1 / Name.synonym: DNA replication factor / Details: UniProtKB/TrEMBL entry Q3MIR3 / Number of copies: 6 / Oligomeric state: hexamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human / Location in cell: nuclear Homo sapiens (human) / synonym: Human / Location in cell: nuclear |
| Molecular weight | Experimental: 590 KDa / Theoretical: 650 KDa |
| Recombinant expression | Organism: Escherichia coli, Rosetta / Recombinant plasmid: pProEX-HT-B |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 6.8 Details: 25 mM Tris-HCl pH 9.0, 150 mM NaCl, 10 mM MgCl2, 50 mM KCl |
| Staining | Type: NEGATIVE Details: Grids were stained with 2% w/v methylamine tungstate, (Nano-W, Nanoprobes Inc.) for 1 min. |
| Grid | Details: 400 mesh, freshly glow-discharged in air |
| Vitrification | Cryogen name: NONE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Electron beam | Acceleration voltage: 100 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.2 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 44000 Bright-field microscopy / Cs: 2.2 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 44000 |
| Sample stage | Specimen holder: eucentric / Specimen holder model: OTHER |
| Details | images were taken on FEI Technai T10 microscope in low dose mode. |
| Date | Jan 1, 2005 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 15 / Average electron dose: 15 e/Å2 / Camera length: 500 / Od range: 1.4 / Bits/pixel: 8 |
- Image processing
Image processing
| CTF correction | Details: each particle |
|---|---|
| Final two d classification | Number classes: 197 |
| Final reconstruction | Applied symmetry - Point group: C6 (6 fold cyclic ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 16.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Imagic / Details: Final map was calculated from 197 best classes / Number images used: 5000 ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 16.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Imagic / Details: Final map was calculated from 197 best classes / Number images used: 5000 |
| Details | The particles were selected interactively at the computer terminal. close |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID -: Chain - Chain ID - 1: L / Chain - Chain ID - 2: T / Chain - Chain ID - 3: L |
|---|---|
| Software | Name: URO |
| Details | Protocol: rigid body. The domains were fitted automatically using URO |
| Refinement | Space: RECIPROCAL / Protocol: RIGID BODY FIT / Target criteria: correlation coeficient |
 Movie
Movie Controller
Controller