[English] 日本語
 Yorodumi
Yorodumi- EMDB-9812: The symmetric-reconstructed cryo-EM structure of Zika virus-FabZK... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9812 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
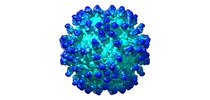
| Title | The symmetric-reconstructed cryo-EM structure of Zika virus-FabZK2B10 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ZIKV / antibody / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to herbicide / photosystem II / photosynthetic electron transport in photosystem II / chlorophyll binding / plasma membrane-derived thylakoid membrane / electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity / metal ion binding Similarity search - Function | |||||||||
| Biological species |   Zika virus / Zika virus /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 11.0 Å | |||||||||
 Authors Authors | Wang L / Wang RK | |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2019 Journal: Cell Rep / Year: 2019Title: Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody. Authors: Lin Wang / Ruoke Wang / Lei Wang / Haijing Ben / Lei Yu / Fei Gao / Xuanling Shi / Chibiao Yin / Fuchun Zhang / Ye Xiang / Linqi Zhang /  Abstract: We previously reported a human monoclonal antibody, ZK2B10, capable of protection against Zika virus (ZIKV) infection and microcephaly in developing mouse embryos. Here, we report the structural ...We previously reported a human monoclonal antibody, ZK2B10, capable of protection against Zika virus (ZIKV) infection and microcephaly in developing mouse embryos. Here, we report the structural features and mechanism of action of ZK2B10. The crystal structure at a resolution of 2.32 Å revealed that the epitope is located on the lateral ridge of DIII of the envelope glycoprotein. Cryo-EM structure with mature ZIKV showed that the antibody binds to DIIIs around the icosahedral 2-fold, 3-fold, and 5-fold axes, a distinct feature compared to those reported for DIII-specific antibodies. The binding of ZK2B10 to ZIKV has no detectable effect on viral attachment to target cells or on conformational changes of the E glycoprotein in the acidic environment, suggesting that ZK2B10 functions at steps between the formation of the fusion intermediate and membrane fusion. These results provide structural and mechanistic insights into how ZK2B10 mediates protection against ZIKV infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9812.map.gz emd_9812.map.gz | 99.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9812-v30.xml emd-9812-v30.xml emd-9812.xml emd-9812.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9812.png emd_9812.png | 266.4 KB | ||
| Filedesc metadata |  emd-9812.cif.gz emd-9812.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9812 http://ftp.pdbj.org/pub/emdb/structures/EMD-9812 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9812 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9812 | HTTPS FTP |
-Validation report
| Summary document |  emd_9812_validation.pdf.gz emd_9812_validation.pdf.gz | 672.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9812_full_validation.pdf.gz emd_9812_full_validation.pdf.gz | 672.2 KB | Display | |
| Data in XML |  emd_9812_validation.xml.gz emd_9812_validation.xml.gz | 7.1 KB | Display | |
| Data in CIF |  emd_9812_validation.cif.gz emd_9812_validation.cif.gz | 8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9812 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9812 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9812 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9812 | HTTPS FTP |
-Related structure data
| Related structure data |  6jfiMC  9811C  6jepC  6jfhC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9812.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9812.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.24 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : ZIKV complexed with FabZK2B10
| Entire | Name: ZIKV complexed with FabZK2B10 |
|---|---|
| Components |
|
-Supramolecule #1: ZIKV complexed with FabZK2B10
| Supramolecule | Name: ZIKV complexed with FabZK2B10 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Zika virus Zika virus |
-Macromolecule #1: ZIKV structural protein E
| Macromolecule | Name: ZIKV structural protein E / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Zika virus Zika virus |
| Molecular weight | Theoretical: 54.186762 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: IRCIGVSNRD FVEGMSGGTW VDVVLEHGGC VTVMAQDKPT VDIELVTTTV SNMAEVRSYC YEASISDMAS DSRCPTQGEA YLDKQSDTQ YVCKRTLVDR GWGNGCGLFG KGSLVTCAKF ACSKKMTGKS IQPENLEYRI MLSVHGSQHS GMIVNDTGHE T DENRAKVE ...String: IRCIGVSNRD FVEGMSGGTW VDVVLEHGGC VTVMAQDKPT VDIELVTTTV SNMAEVRSYC YEASISDMAS DSRCPTQGEA YLDKQSDTQ YVCKRTLVDR GWGNGCGLFG KGSLVTCAKF ACSKKMTGKS IQPENLEYRI MLSVHGSQHS GMIVNDTGHE T DENRAKVE ITPNSPRAEA TLGGFGSLGL DCEPRTGLDF SDLYYLTMNN KHWLVHKEWF HDIPLPWHAG ADTGTPHWNN KE ALVEFKD AHAKRQTVVV LGSQEGAVHT ALAGALEAEM DGAKGRLSSG HLKCRLKMDK LRLKGVSYSL CTAAFTFTKI PAE TLHGTV TVEVQYAGTD GPCKVPAQMA VDMQTLTPVG RLITANPVIT ESTENSKMML ELDPPFGDSY IVIGVGEKKI THHW HRSGS TIGKAFEATV RGAKRMAVLG DTAWDFGSVG GALNSLGKGI HQIFGAAFKS LFGGMSWFSQ ILIGTLLMWL GLNTK NGSI SLMCLALGGV LIFLSTA |
-Macromolecule #2: strutural protein M
| Macromolecule | Name: strutural protein M / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO / EC number: flavivirin |
|---|---|
| Source (natural) | Organism:   Zika virus Zika virus |
| Molecular weight | Theoretical: 8.496883 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVTLPSHSTR KLQTRSQTWL ESREYTKHLI RVENWIFRNP GFALAAAAIA WLLGSSTSQK VIYLVMILLI APAYS UniProtKB: Photosystem q(B) protein |
-Macromolecule #3: FabZK2B10 heavy chain
| Macromolecule | Name: FabZK2B10 heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.383305 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVQSGAE VKKPGASVKV SCKASGYTFT SGHTFPSYDI NWVRQATGQG LEWMGWMNPN RGNTGYAQKF QGRVTMTRNT SINTAYMEL SSLRSEDTAV YYCARVRSGT NYGSYYYYYY GMDVWGQGTT VTVSSASTKG PSVFPLAPSS KSTSGGTAAL G CLVKDYFP ...String: EVQLVQSGAE VKKPGASVKV SCKASGYTFT SGHTFPSYDI NWVRQATGQG LEWMGWMNPN RGNTGYAQKF QGRVTMTRNT SINTAYMEL SSLRSEDTAV YYCARVRSGT NYGSYYYYYY GMDVWGQGTT VTVSSASTKG PSVFPLAPSS KSTSGGTAAL G CLVKDYFP EPVTVSWNSG ALTSGVHTFP AVLQSSGLYS LSSVVTVPSS SLGTQTYICN VNHKPSNTKV DKRVEPK |
-Macromolecule #4: FabZK2B10 light chain
| Macromolecule | Name: FabZK2B10 light chain / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.782035 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SYELTQPPSA SGTPGQRVTI SCSGSSSNIG NNYVHWYQQL PGSAPKLLIY RNNQRPSGVP DRFSGSKSGT SGSLAISGLR SEDEADYYC ASWDDSLSGH WVFGGGTKVT VLGQPKAAPS VTLFPPSSEE LQANKATLVC LISDFYPGAV TVAWKADSSP V KAGVETTT ...String: SYELTQPPSA SGTPGQRVTI SCSGSSSNIG NNYVHWYQQL PGSAPKLLIY RNNQRPSGVP DRFSGSKSGT SGSLAISGLR SEDEADYYC ASWDDSLSGH WVFGGGTKVT VLGQPKAAPS VTLFPPSSEE LQANKATLVC LISDFYPGAV TVAWKADSSP V KAGVETTT PSKQSNNKYA ASSYLSLTPE QWKSHRSYSC QVTHEGSTVE KTVAPT |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Resolution.type: BY AUTHOR / Resolution: 11.0 Å / Resolution method: OTHER / Number images used: 684 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Initial model |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Output model |  PDB-6jfi: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)