[English] 日本語
 Yorodumi
Yorodumi- EMDB-9665: The cryoEM map of HPV33 VLP in complex with the Fab fragment of a... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9665 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryoEM map of HPV33 VLP in complex with the Fab fragment of antibody 4E5 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Human papillomavirus type 33 Human papillomavirus type 33 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 12.3 Å | |||||||||
 Authors Authors | Li ZH / Song S / He MZ / Gu Y / Li SW | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity. Authors: Zhihai Li / Shuo Song / Maozhou He / Daning Wang / Jingjie Shi / Xinlin Liu / Yunbing Li / Xin Chi / Shuangping Wei / Yurou Yang / Zhiping Wang / Jinjin Li / Huilian Qian / Hai Yu / Qingbing ...Authors: Zhihai Li / Shuo Song / Maozhou He / Daning Wang / Jingjie Shi / Xinlin Liu / Yunbing Li / Xin Chi / Shuangping Wei / Yurou Yang / Zhiping Wang / Jinjin Li / Huilian Qian / Hai Yu / Qingbing Zheng / Xiaodong Yan / Qinjian Zhao / Jun Zhang / Ying Gu / Shaowei Li / Ningshao Xia /   Abstract: Sequence variability in surface-antigenic sites of pathogenic proteins is an important obstacle in vaccine development. Over 200 distinct genomic sequences have been identified for human ...Sequence variability in surface-antigenic sites of pathogenic proteins is an important obstacle in vaccine development. Over 200 distinct genomic sequences have been identified for human papillomavirus (HPV), of which more than 18 are associated with cervical cancer. Here, based on the high structural similarity of L1 surface loops within a group of phylogenetically close HPV types, we design a triple-type chimera of HPV33/58/52 using loop swapping. The chimeric VLPs elicit neutralization titers comparable with a mix of the three wild-type VLPs both in mice and non-human primates. This engineered region of the chimeric protein recapitulates the conformational contours of the antigenic surfaces of the parental-type proteins, offering a basis for this high immunity. Our stratagem is equally successful in developing other triplet-type chimeras (HPV16/35/31, HPV56/66/53, HPV39/68/70, HPV18/45/59), paving the way for the development of an improved HPV prophylactic vaccine against all carcinogenic HPV strains. This technique may also be extrapolated to other microbes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9665.map.gz emd_9665.map.gz | 884 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9665-v30.xml emd-9665-v30.xml emd-9665.xml emd-9665.xml | 8.3 KB 8.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9665.png emd_9665.png | 104.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9665 http://ftp.pdbj.org/pub/emdb/structures/EMD-9665 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9665 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9665 | HTTPS FTP |
-Validation report
| Summary document |  emd_9665_validation.pdf.gz emd_9665_validation.pdf.gz | 78.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9665_full_validation.pdf.gz emd_9665_full_validation.pdf.gz | 77.3 KB | Display | |
| Data in XML |  emd_9665_validation.xml.gz emd_9665_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9665 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9665 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9665 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9665 | HTTPS FTP |
-Related structure data
| Related structure data |  9666C  9667C  6igcC  6igdC  6igeC  6igfC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9665.map.gz / Format: CCP4 / Size: 2.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9665.map.gz / Format: CCP4 / Size: 2.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.128 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
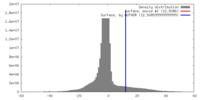
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human papillomavirus type 33
| Entire | Name:  Human papillomavirus type 33 Human papillomavirus type 33 |
|---|---|
| Components |
|
-Supramolecule #1: Human papillomavirus type 33
| Supramolecule | Name: Human papillomavirus type 33 / type: virus / ID: 1 / Parent: 0 Details: VLP generated by recombinantly expressed; Fab Fragment generated by proteolytic cleavage of IgG antibody; NCBI-ID: 10586 / Sci species name: Human papillomavirus type 33 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host system | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 12.3 Å / Resolution method: OTHER / Details: FSC 0.3 cut-off / Number images used: 753 |
|---|---|
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: COMMON LINE |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)