[English] 日本語
 Yorodumi
Yorodumi- EMDB-9352: Cryo-EM structure of singly-bound SNF2h-nucleosome complex at 3.4 A -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9352 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of singly-bound SNF2h-nucleosome complex at 3.4 A | ||||||||||||||||||
 Map data Map data | Cryosparc v2 Non-Uniform refinement map. bfactor -130; Resampled into a position to fit with other maps uploaded | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Biological species | |||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.39 Å | ||||||||||||||||||
 Authors Authors | Armache J-P / Gamarra N / Johnson SL / Wu S / Leonard JD / Narlikar GJ / Cheng Y | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome. Authors: Jean Paul Armache / Nathan Gamarra / Stephanie L Johnson / John D Leonard / Shenping Wu / Geeta J Narlikar / Yifan Cheng /  Abstract: The SNF2h remodeler slides nucleosomes most efficiently as a dimer, yet how the two protomers avoid a tug-of-war is unclear. Furthermore, SNF2h couples histone octamer deformation to nucleosome ...The SNF2h remodeler slides nucleosomes most efficiently as a dimer, yet how the two protomers avoid a tug-of-war is unclear. Furthermore, SNF2h couples histone octamer deformation to nucleosome sliding, but the underlying structural basis remains unknown. Here we present cryo-EM structures of SNF2h-nucleosome complexes with ADP-BeF that capture two potential reaction intermediates. In one structure, histone residues near the dyad and in the H2A-H2B acidic patch, distal to the active SNF2h protomer, appear disordered. The disordered acidic patch is expected to inhibit the second SNF2h protomer, while disorder near the dyad is expected to promote DNA translocation. The other structure doesn't show octamer deformation, but surprisingly shows a 2 bp translocation. FRET studies indicate that ADP-BeF predisposes SNF2h-nucleosome complexes for an elemental translocation step. We propose a model for allosteric control through the nucleosome, where one SNF2h protomer promotes asymmetric octamer deformation to inhibit the second protomer, while stimulating directional DNA translocation. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9352.map.gz emd_9352.map.gz | 74.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9352-v30.xml emd-9352-v30.xml emd-9352.xml emd-9352.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9352.png emd_9352.png | 207.8 KB | ||
| Others |  emd_9352_additional.map.gz emd_9352_additional.map.gz emd_9352_half_map_1.map.gz emd_9352_half_map_1.map.gz emd_9352_half_map_2.map.gz emd_9352_half_map_2.map.gz | 51.9 MB 95.4 MB 95.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9352 http://ftp.pdbj.org/pub/emdb/structures/EMD-9352 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9352 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9352 | HTTPS FTP |
-Validation report
| Summary document |  emd_9352_validation.pdf.gz emd_9352_validation.pdf.gz | 78.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9352_full_validation.pdf.gz emd_9352_full_validation.pdf.gz | 77.8 KB | Display | |
| Data in XML |  emd_9352_validation.xml.gz emd_9352_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9352 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9352 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9352 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9352 | HTTPS FTP |
-Related structure data
| Related structure data |  9351C  9353C  9354C  9355C  9356C  6ne3C C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10287 (Title: Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome EMPIAR-10287 (Title: Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosomeData size: 1.4 TB Data #1: Unaligned multi-frame micrographs of SNF2h bound to a nucleosome [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9352.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9352.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryosparc v2 Non-Uniform refinement map. bfactor -130; Resampled into a position to fit with other maps uploaded | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2156 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Cryosparc v2 Non-Uniform refinement map. No bfactor; In...
| File | emd_9352_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryosparc v2 Non-Uniform refinement map. No bfactor; In original position | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryosparc v2 Non-Uniform refinement half-map 2. No bfactor;...
| File | emd_9352_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryosparc v2 Non-Uniform refinement half-map 2. No bfactor; In original position | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryosparc v2 Non-Uniform refinement half-map 1. No bfactor;...
| File | emd_9352_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryosparc v2 Non-Uniform refinement half-map 1. No bfactor; In original position | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of singly-bound SNF2h-nucleosome complex at 3.4 A
| Entire | Name: Cryo-EM structure of singly-bound SNF2h-nucleosome complex at 3.4 A |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of singly-bound SNF2h-nucleosome complex at 3.4 A
| Supramolecule | Name: Cryo-EM structure of singly-bound SNF2h-nucleosome complex at 3.4 A type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism: |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 340 kDa/nm |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295.15 K / Instrument: FEI VITROBOT MARK I Details: 2.5 ul of nucleosome-443 SNF2h complexes were applied to a glow discharged Quantifoil holey carbon grid (1.2 um hole size, 400 mesh), blotted in a Vitrobot Mark I (FEI Company) using 6 ...Details: 2.5 ul of nucleosome-443 SNF2h complexes were applied to a glow discharged Quantifoil holey carbon grid (1.2 um hole size, 400 mesh), blotted in a Vitrobot Mark I (FEI Company) using 6 seconds blotting at 100% humidity, and then plunge-frozen in liquid ethane cooled by liquid nitrogen.. |
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 41.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|
 Movie
Movie Controller
Controller


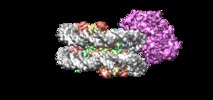
 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)