[English] 日本語
 Yorodumi
Yorodumi- EMDB-9070: Single-Molecule 3D Image of Human Plasma Intermediate-Density Lip... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9070 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single-Molecule 3D Image of Human Plasma Intermediate-Density Lipoprotein (No. 02) | |||||||||||||||
 Map data Map data | primary map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
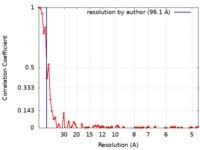
| Method | electron tomography / cryo EM / Resolution: 96.1 Å | |||||||||||||||
 Authors Authors | Lei D / Yu Y / Kuang Y / Krauss R / Ren G | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Biochim Biophys Acta Mol Cell Biol Lipids / Year: 2019 Journal: Biochim Biophys Acta Mol Cell Biol Lipids / Year: 2019Title: Single-molecule 3D imaging of human plasma intermediate-density lipoproteins reveals a polyhedral structure. Authors: Dongsheng Lei / Yadong Yu / Yu-Lin Kuang / Jianfang Liu / Ronald M Krauss / Gang Ren /  Abstract: Intermediate-density lipoproteins (IDLs), the remnants of very-low-density lipoproteins via lipolysis, are rich in cholesteryl ester and are associated with cardiovascular disease. Despite ...Intermediate-density lipoproteins (IDLs), the remnants of very-low-density lipoproteins via lipolysis, are rich in cholesteryl ester and are associated with cardiovascular disease. Despite pharmacological interest in IDLs, their three-dimensional (3D) structure is still undetermined due to their variation in size, composition, and dynamic structure. To explore the 3D structure of IDLs, we reconstructed 3D density maps from individual IDL particles using cryo-electron microscopy (cryo-EM) and individual-particle electron tomography (IPET, without averaging from different molecules). 3D reconstructions of IDLs revealed an unexpected polyhedral structure that deviates from the generally assumed spherical shape model (Frias et al., 2007; Olson, 1998; Shen et al., 1977). The polyhedral-shaped IDL contains a high-density shell formed by flat surfaces that are similar to those of very-low-density lipoproteins but have sharper dihedral angles between nearby surfaces. These flat surfaces would be less hydrophobic than the curved surface of mature spherical high-density lipoprotein (HDL), leading to a lower binding affinity of IDL to hydrophobic proteins (such as cholesteryl ester transfer protein) than HDL. This is the first visualization of the IDL 3D structure, which could provide fundamental clues for delineating the role of IDL in lipid metabolism and cardiovascular disease. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9070.map.gz emd_9070.map.gz | 24.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9070-v30.xml emd-9070-v30.xml emd-9070.xml emd-9070.xml | 10.4 KB 10.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9070_fsc.xml emd_9070_fsc.xml | 6.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_9070.png emd_9070.png | 22.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9070 http://ftp.pdbj.org/pub/emdb/structures/EMD-9070 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9070 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9070 | HTTPS FTP |
-Validation report
| Summary document |  emd_9070_validation.pdf.gz emd_9070_validation.pdf.gz | 78.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9070_full_validation.pdf.gz emd_9070_full_validation.pdf.gz | 77.8 KB | Display | |
| Data in XML |  emd_9070_validation.xml.gz emd_9070_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9070 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9070 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9070 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9070 | HTTPS FTP |
-Related structure data
| Related structure data |  9069C  9071C  9072C  9073C  9074C  9075C  9076C  9077C  9078C  9079C  9080C  9081C  9082C  9083C  9084C  9085C  9086C  9087C  9088C  9089C  9090C  9091C 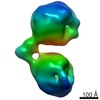 9092C  9093C  9094C 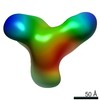 9095C  9096C 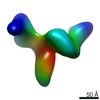 9097C 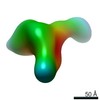 9098C  9099C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9070.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9070.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | primary map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human plasma intermediate-density lipoprotein
| Entire | Name: Human plasma intermediate-density lipoprotein |
|---|---|
| Components |
|
-Supramolecule #1: Human plasma intermediate-density lipoprotein
| Supramolecule | Name: Human plasma intermediate-density lipoprotein / type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: Blood Homo sapiens (human) / Tissue: Blood |
| Molecular weight | Experimental: 3 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7 / Details: Dulbecco's phosphate buffered saline (DPBS) |
| Grid | Model: Homemade / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Instrument: LEICA EM GP |
| Details | Human IDLs were isolated from the plasma of a healthy individual by non-equilibrium density gradient ultracentrifugation |
| Sectioning | Other: NO SECTIONING |
- Electron microscopy
Electron microscopy
| Microscope | ZEISS LIBRA120PLUS |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average electron dose: 1.45 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal magnification: 50000 |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera

 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)