+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8912 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Polycsytin 2-l1 | |||||||||
 Map data Map data | Human Polycsytin 2-l1, primary map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationsour taste receptor activity / detection of chemical stimulus involved in sensory perception of sour taste / detection of chemical stimulus involved in sensory perception of taste / sensory perception of sour taste / response to water / calcium-activated potassium channel activity / detection of mechanical stimulus / muscle alpha-actinin binding / cellular response to acidic pH / calcium-activated cation channel activity ...sour taste receptor activity / detection of chemical stimulus involved in sensory perception of sour taste / detection of chemical stimulus involved in sensory perception of taste / sensory perception of sour taste / response to water / calcium-activated potassium channel activity / detection of mechanical stimulus / muscle alpha-actinin binding / cellular response to acidic pH / calcium-activated cation channel activity / sodium channel activity / non-motile cilium / inorganic cation transmembrane transport / ciliary membrane / smoothened signaling pathway / alpha-actinin binding / monoatomic cation transport / sodium ion transmembrane transport / monoatomic cation channel activity / potassium ion transmembrane transport / calcium channel complex / cytoskeletal protein binding / calcium channel activity / actin cytoskeleton / cytoplasmic vesicle / protein homotetramerization / transmembrane transporter binding / receptor complex / intracellular membrane-bounded organelle / calcium ion binding / cell surface / endoplasmic reticulum / membrane / identical protein binding / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
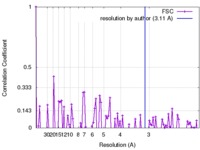
| Method | single particle reconstruction / cryo EM / Resolution: 3.11 Å | |||||||||
 Authors Authors | Hulse RE / Clapham DE / Li Z / Huang RK / Zhang J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2018 Journal: Elife / Year: 2018Title: Cryo-EM structure of the polycystin 2-l1 ion channel. Authors: Raymond E Hulse / Zongli Li / Rick K Huang / Jin Zhang / David E Clapham /   Abstract: We report the near atomic resolution (3.3 Å) of the human polycystic kidney disease 2-like 1 (polycystin 2-l1) ion channel. Encoded by PKD2L1, polycystin 2-l1 is a calcium and monovalent cation- ...We report the near atomic resolution (3.3 Å) of the human polycystic kidney disease 2-like 1 (polycystin 2-l1) ion channel. Encoded by PKD2L1, polycystin 2-l1 is a calcium and monovalent cation-permeant ion channel in primary cilia and plasma membranes. The related primary cilium-specific polycystin-2 protein, encoded by PKD2, shares a high degree of sequence similarity, yet has distinct permeability characteristics. Here we show that these differences are reflected in the architecture of polycystin 2-l1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8912.map.gz emd_8912.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8912-v30.xml emd-8912-v30.xml emd-8912.xml emd-8912.xml | 12.4 KB 12.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8912_fsc.xml emd_8912_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_8912.png emd_8912.png | 3.3 MB | ||
| Masks |  emd_8912_msk_1.map emd_8912_msk_1.map | 64 MB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8912 http://ftp.pdbj.org/pub/emdb/structures/EMD-8912 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8912 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8912 | HTTPS FTP |
-Validation report
| Summary document |  emd_8912_validation.pdf.gz emd_8912_validation.pdf.gz | 592.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8912_full_validation.pdf.gz emd_8912_full_validation.pdf.gz | 591.9 KB | Display | |
| Data in XML |  emd_8912_validation.xml.gz emd_8912_validation.xml.gz | 11 KB | Display | |
| Data in CIF |  emd_8912_validation.cif.gz emd_8912_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8912 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8912 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8912 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8912 | HTTPS FTP |
-Related structure data
| Related structure data |  6du8MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8912.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8912.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human Polycsytin 2-l1, primary map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
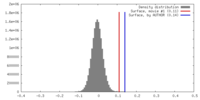
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_8912_msk_1.map emd_8912_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
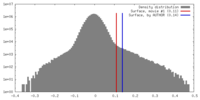
| Density Histograms |
- Sample components
Sample components
-Entire : Human polycystic 2-l
| Entire | Name: Human polycystic 2-l |
|---|---|
| Components |
|
-Supramolecule #1: Human polycystic 2-l
| Supramolecule | Name: Human polycystic 2-l / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: Homotetrameric assembly of polycystic 2-l1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 547 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant plasmid: pEG Homo sapiens (human) / Recombinant plasmid: pEG |
-Macromolecule #1: Polycystic kidney disease 2-like 1 protein
| Macromolecule | Name: Polycystic kidney disease 2-like 1 protein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 92.070633 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MNAVGSPEGQ ELQKLGSGAW DNPAYSGPPS PHGTLRVCTI SSTGPLQPQP KKPEDEPQET AYRTQVSSCC LHICQGIRGL WGTTLTENT AENRELYIKT TLRELLVYIV FLVDICLLTY GMTSSSAYYY TKVMSELFLH TPSDTGVSFQ AISSMADFWD F AQGPLLDS ...String: MNAVGSPEGQ ELQKLGSGAW DNPAYSGPPS PHGTLRVCTI SSTGPLQPQP KKPEDEPQET AYRTQVSSCC LHICQGIRGL WGTTLTENT AENRELYIKT TLRELLVYIV FLVDICLLTY GMTSSSAYYY TKVMSELFLH TPSDTGVSFQ AISSMADFWD F AQGPLLDS LYWTKWYNNQ SLGHGSHSFI YYENMLLGVP RLRQLKVRND SCVVHEDFRE DILSCYDVYS PDKEEQLPFG PF NGTAWTY HSQDELGGFS HWGRLTSYSG GGYYLDLPGS RQGSAEALRA LQEGLWLDRG TRVVFIDFSV YNANINLFCV LRL VVEFPA TGGAIPSWQI RTVKLIRYVS NWDFFIVGCE VIFCVFIFYY VVEEILELHI HRLRYLSSIW NILDLVVILL SIVA VGFHI FRTLEVNRLM GKLLQQPNTY ADFEFLAFWQ TQYNNMNAVN LFFAWIKIFK YISFNKTMTQ LSSTLARCAK DILGF AVMF FIVFFAYAQL GYLLFGTQVE NFSTFIKCIF TQFRIILGDF DYNAIDNANR ILGPAYFVTY VFFVFFVLLN MFLAII NDT YSEVKEELAG QKDELQLSDL LKQGYNKTLL RLRLRKERVS DVQKVLQGGE QEIQFEDFTN TLRELGHAEH EITELTA TF TKFDRDGNRI LDEKEQEKMR QDLEEERVAL NTEIEKLGRS IVSSPQGKSG PEAARAGGWV SGEEFYMLTR RVLQLETV L EGVVSQIDAV GSKLKMLERK GWLAPSPGVK EQAIWKHPQP APAVTPDPWG VQGGQESEVP YKREEEALEE RRLSRGEIP TLQRS |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Concentration | 3.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: FORMVAR / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | polycystic 2-l1 stabilized in amphipol PMAL-C8 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)