[English] 日本語
 Yorodumi
Yorodumi- EMDB-8792: MERS S ectodomain trimer in complex with Fab of neutralizing anti... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8792 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | MERS S ectodomain trimer in complex with Fab of neutralizing antibody G4 | |||||||||
 Map data Map data | MERS S ectodomain trimer in complex with variable domain of neutralizing antibody G4 | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationmembrane fusion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
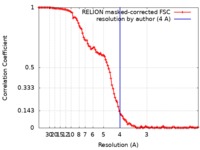
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Pallesen J / Ward AB | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2017 Journal: Proc Natl Acad Sci U S A / Year: 2017Title: Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Authors: Jesper Pallesen / Nianshuang Wang / Kizzmekia S Corbett / Daniel Wrapp / Robert N Kirchdoerfer / Hannah L Turner / Christopher A Cottrell / Michelle M Becker / Lingshu Wang / Wei Shi / Wing- ...Authors: Jesper Pallesen / Nianshuang Wang / Kizzmekia S Corbett / Daniel Wrapp / Robert N Kirchdoerfer / Hannah L Turner / Christopher A Cottrell / Michelle M Becker / Lingshu Wang / Wei Shi / Wing-Pui Kong / Erica L Andres / Arminja N Kettenbach / Mark R Denison / James D Chappell / Barney S Graham / Andrew B Ward / Jason S McLellan /  Abstract: Middle East respiratory syndrome coronavirus (MERS-CoV) is a lineage C betacoronavirus that since its emergence in 2012 has caused outbreaks in human populations with case-fatality rates of ∼36%. ...Middle East respiratory syndrome coronavirus (MERS-CoV) is a lineage C betacoronavirus that since its emergence in 2012 has caused outbreaks in human populations with case-fatality rates of ∼36%. As in other coronaviruses, the spike (S) glycoprotein of MERS-CoV mediates receptor recognition and membrane fusion and is the primary target of the humoral immune response during infection. Here we use structure-based design to develop a generalizable strategy for retaining coronavirus S proteins in the antigenically optimal prefusion conformation and demonstrate that our engineered immunogen is able to elicit high neutralizing antibody titers against MERS-CoV. We also determined high-resolution structures of the trimeric MERS-CoV S ectodomain in complex with G4, a stem-directed neutralizing antibody. The structures reveal that G4 recognizes a glycosylated loop that is variable among coronaviruses and they define four conformational states of the trimer wherein each receptor-binding domain is either tightly packed at the membrane-distal apex or rotated into a receptor-accessible conformation. Our studies suggest a potential mechanism for fusion initiation through sequential receptor-binding events and provide a foundation for the structure-based design of coronavirus vaccines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8792.map.gz emd_8792.map.gz | 99.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8792-v30.xml emd-8792-v30.xml emd-8792.xml emd-8792.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8792_fsc.xml emd_8792_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_8792.png emd_8792.png | 86 KB | ||
| Others |  emd_8792_half_map_1.map.gz emd_8792_half_map_1.map.gz emd_8792_half_map_2.map.gz emd_8792_half_map_2.map.gz | 84.3 MB 84.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8792 http://ftp.pdbj.org/pub/emdb/structures/EMD-8792 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8792 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8792 | HTTPS FTP |
-Validation report
| Summary document |  emd_8792_validation.pdf.gz emd_8792_validation.pdf.gz | 78.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8792_full_validation.pdf.gz emd_8792_full_validation.pdf.gz | 77.6 KB | Display | |
| Data in XML |  emd_8792_validation.xml.gz emd_8792_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8792 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8792 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8792 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8792 | HTTPS FTP |
-Related structure data
| Related structure data |  8783C  8784C  8785C  8786C  8787C  8788C  8789C  8790C  8791C  8793C  5vyhC  5vzrC  5w9hC  5w9iC  5w9jC  5w9kC  5w9lC  5w9mC  5w9nC  5w9oC  5w9pC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8792.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8792.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MERS S ectodomain trimer in complex with variable domain of neutralizing antibody G4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.02 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: MERS S ectodomain trimer in complex with variable...
| File | emd_8792_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MERS S ectodomain trimer in complex with variable domain of neutralizing antibody G4 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: MERS S ectodomain trimer in complex with variable...
| File | emd_8792_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MERS S ectodomain trimer in complex with variable domain of neutralizing antibody G4 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MERS S ectodomain trimer in complex with Fab of neutralizing anti...
| Entire | Name: MERS S ectodomain trimer in complex with Fab of neutralizing antibody G4 |
|---|---|
| Components |
|
-Supramolecule #1: MERS S ectodomain trimer in complex with Fab of neutralizing anti...
| Supramolecule | Name: MERS S ectodomain trimer in complex with Fab of neutralizing antibody G4 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 600 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Name: TBS |
| Grid | Model: C-flat / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Sampling interval: 5.0 µm / Digitization - Frames/image: 1-35 / Number grids imaged: 1 / Average exposure time: 0.2 sec. / Average electron dose: 1.89 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)