[English] 日本語
 Yorodumi
Yorodumi- EMDB-6396: The structure of BipA in GTP form bound to the ratcheted ribosome -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6396 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of BipA in GTP form bound to the ratcheted ribosome | |||||||||
 Map data Map data | Reconstruction of BipA bound to 70S | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribosome / BipA | |||||||||
| Function / homology |  Function and homology information Function and homology informationguanosine tetraphosphate binding / protein folding chaperone / response to cold / large ribosomal subunit / ribosome binding / regulation of translation / ribosomal small subunit assembly / small ribosomal subunit / 5S rRNA binding / large ribosomal subunit rRNA binding ...guanosine tetraphosphate binding / protein folding chaperone / response to cold / large ribosomal subunit / ribosome binding / regulation of translation / ribosomal small subunit assembly / small ribosomal subunit / 5S rRNA binding / large ribosomal subunit rRNA binding / transferase activity / response to heat / cytosolic small ribosomal subunit / ribosomal large subunit assembly / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / cytosolic large ribosomal subunit / tRNA binding / negative regulation of translation / rRNA binding / ribosome / structural constituent of ribosome / ribonucleoprotein complex / translation / GTPase activity / mRNA binding / GTP binding / zinc ion binding / metal ion binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.7 Å | |||||||||
 Authors Authors | Kumar V / Chen Y / Ero R / Ahmed T / Tan J / Li Z / Wong ASW / Gao Y-G / Bhushan S | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2015 Journal: Proc Natl Acad Sci U S A / Year: 2015Title: Structure of BipA in GTP form bound to the ratcheted ribosome. Authors: Veerendra Kumar / Yun Chen / Rya Ero / Tofayel Ahmed / Jackie Tan / Zhe Li / Andrew See Weng Wong / Shashi Bhushan / Yong-Gui Gao /  Abstract: BPI-inducible protein A (BipA) is a member of the family of ribosome-dependent translational GTPase (trGTPase) factors along with elongation factors G and 4 (EF-G and EF4). Despite being highly ...BPI-inducible protein A (BipA) is a member of the family of ribosome-dependent translational GTPase (trGTPase) factors along with elongation factors G and 4 (EF-G and EF4). Despite being highly conserved in bacteria and playing a critical role in coordinating cellular responses to environmental changes, its structures (isolated and ribosome bound) remain elusive. Here, we present the crystal structures of apo form and GTP analog, GDP, and guanosine-3',5'-bisdiphosphate (ppGpp)-bound BipA. In addition to having a distinctive domain arrangement, the C-terminal domain of BipA has a unique fold. Furthermore, we report the cryo-electron microscopy structure of BipA bound to the ribosome in its active GTP form and elucidate the unique structural attributes of BipA interactions with the ribosome and A-site tRNA in the light of its possible function in regulating translation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6396.map.gz emd_6396.map.gz | 4.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6396-v30.xml emd-6396-v30.xml emd-6396.xml emd-6396.xml | 9.9 KB 9.9 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6396.gif 400_6396.gif 80_6396.gif 80_6396.gif | 76.8 KB 4.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6396 http://ftp.pdbj.org/pub/emdb/structures/EMD-6396 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6396 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6396 | HTTPS FTP |
-Validation report
| Summary document |  emd_6396_validation.pdf.gz emd_6396_validation.pdf.gz | 253.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6396_full_validation.pdf.gz emd_6396_full_validation.pdf.gz | 252.4 KB | Display | |
| Data in XML |  emd_6396_validation.xml.gz emd_6396_validation.xml.gz | 6.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6396 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6396 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6396 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6396 | HTTPS FTP |
-Related structure data
| Related structure data |  5a9zMC  6397C  5a9vC  5a9wC  5a9xC  5a9yC  5aa0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6396.map.gz / Format: CCP4 / Size: 94.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6396.map.gz / Format: CCP4 / Size: 94.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of BipA bound to 70S | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.24 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
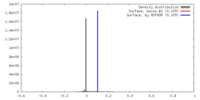
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : BipA bound to 70S
| Entire | Name: BipA bound to 70S |
|---|---|
| Components |
|
-Supramolecule #1000: BipA bound to 70S
| Supramolecule | Name: BipA bound to 70S / type: sample / ID: 1000 / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 2.5 MDa / Theoretical: 2.5 MDa / Method: Sedimentation |
-Supramolecule #1: 70S ribosome
| Supramolecule | Name: 70S ribosome / type: complex / ID: 1 / Recombinant expression: No / Database: NCBI Ribosome-details: ribosome-prokaryote: LSU 50S, LSU RNA 23S, LSU RNA 5S, SSU 30S, PSR16s, ALL |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| Molecular weight | Experimental: 2.5 MDa / Theoretical: 2.5 MDa |
-Macromolecule #1: BipA
| Macromolecule | Name: BipA / type: protein_or_peptide / ID: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: 5 mM HEPES, pH 7.5, 10 mM MgOAc, 50 mM KCl, 10 mM NH4Cl, 6 mM 2-mercaptoethanol |
|---|---|
| Grid | Details: 300 mesh copper grid with 2 nm carbon support, glow-discharged |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV / Method: Blot for 3 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 73684 times magnification. |
| Date | Jan 27, 2015 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 658 / Average electron dose: 20 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 73684 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 53000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Particles were selected using EMAN and processed using Relion. |
|---|---|
| CTF correction | Details: CTFfind |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 4.7 Å / Resolution method: OTHER / Software - Name: Relion / Number images used: 61165 |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)